Hello,
I used the documentation of running single point configuration for Alaska with CLM5 CESM 2.1.3 and the case I used has --res f19_g16 --compset I2000Clm50BgcCruGs and runs with my own machine config (that is tested and has worked with other compset).
I used this link info to make the case and after it downloaded all of the required input data I submitted the case successfully.
I have to mention that in some parts I had to manually download some domain and surface_map data using the following command:
wget https://svn-ccsm-inputdata.cgd.ucar...clm/domain.lnd.13x12pt_f19_alaskaUSA_gx1v6.nc -P /home/XXXXX/projects/YYYYY/XXXXX/inputdata/share/domains/ --no-check-certificate
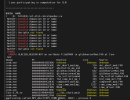
But after submission, it failed with 1 min of running with the following message which appears in the log file multiple times:
MPI_ABORT was invoked on rank 55 in communicator MPI_COMM_WORLD with errorcode 1001. NOTE: invoking MPI_ABORT causes Open MPI to kill all MPI processes. You may or may not see output from other processes, depending on exactly when Open MPI kills them.
I searched the forum and couldn't find the reason why I am having this error. I appreciate it if you let me know your opinion.
I used the documentation of running single point configuration for Alaska with CLM5 CESM 2.1.3 and the case I used has --res f19_g16 --compset I2000Clm50BgcCruGs and runs with my own machine config (that is tested and has worked with other compset).
I used this link info to make the case and after it downloaded all of the required input data I submitted the case successfully.
I have to mention that in some parts I had to manually download some domain and surface_map data using the following command:
wget https://svn-ccsm-inputdata.cgd.ucar...clm/domain.lnd.13x12pt_f19_alaskaUSA_gx1v6.nc -P /home/XXXXX/projects/YYYYY/XXXXX/inputdata/share/domains/ --no-check-certificate
But after submission, it failed with 1 min of running with the following message which appears in the log file multiple times:
MPI_ABORT was invoked on rank 55 in communicator MPI_COMM_WORLD with errorcode 1001. NOTE: invoking MPI_ABORT causes Open MPI to kill all MPI processes. You may or may not see output from other processes, depending on exactly when Open MPI kills them.
I searched the forum and couldn't find the reason why I am having this error. I appreciate it if you let me know your opinion.