I am trying to run the model for a single year using the 2000_DATM%GSWP3v1_CLM50%SP_SICE_SOCN_MOSART_SGLC_SWAV compset. The region I am focusing on is the Central U.S. (latitude 33–46°, longitude 250–272°), and I am using a high spatial resolution of 0.05 degrees for the atmospheric forcing data.
For the surface dataset, I used:
/glade/campaign/cgd/tss/people/oleson/CLM5_datasets/ctsm5.2.0/GLOBAL/surfdata_0.05x0.05-hires_hist_2005_78pfts_c240425.nc
because I want to run the model at a fine resolution. Since the monthly LAI data in the surface dataset follows the same annual cycle each year, I initially enabled the use of external LAI by setting use_lai_streams = .true. in user_nl_clm.
However, after submitting the case, I encountered an error.
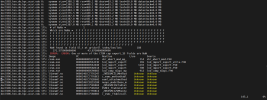
/glade/derecho/scratch/koushanm/April_30_USA_MODIS_SP/run> vi cesm.log.9370208.desched1.250430-213018
dec1906.hsn.de.hpc.ucar.edu 175: # of NaNs = 1
dec1906.hsn.de.hpc.ucar.edu 175: Which are NaNs = F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F T F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: NaN found in field Sl_t at gridcell index/lon/lat: 188
dec1906.hsn.de.hpc.ucar.edu 175: 271.575000000000 43.8750000000000
dec1906.hsn.de.hpc.ucar.edu 175: ERROR: ERROR: One or more of the CTSM cap export_1D fields are NaN
dec1906.hsn.de.hpc.ucar.edu 175: Image PC Routine Line Source
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 000000000107373D shr_abort_mod_mp_ 114 shr_abort_mod.F90
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 00000000005A3E3D lnd_import_export 169 lnd_import_export_utils.F90
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 00000000005A3040 lnd_import_export 1193 lnd_import_export.F90
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 00000000005A0618 lnd_import_export 780 lnd_import_export.F90
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 0000000000590083 lnd_comp_nuopc_mp 912 lnd_comp_nuopc.F90
In the next step, I attempted to crop the MODIS LAI stream data using the following command:
ncks -O -d lat,33,46 -d lon,250,272 /glade/u/home/koushanm/MODISPFTLAI_0.5x0.5_c140711.nc MODISPFTLAI_cropped.nc
I also used a mesh file created based on the surface dataset and added the following settings to user_nl_clm:
stream_fldfilename_lai = '/glade/u/home/koushanm/MODISPFTLAI_cropped.nc'
stream_meshfile_lai = '/glade/work/koushanm/ctsm5.2.005/CTSM/tools/site_and_regional/subset_data_regional/lnd_mesh.nc'
After submitting the case again, I encountered another error. The new log file is located at:
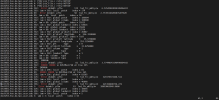
/glade/derecho/scratch/koushanm/April_30_USA_MODIS_SP/run/cesm.log.9370835.desched1.250430-222056
dec0253.hsn.de.hpc.ucar.edu 0: PIO2 pio_file.c retry NETCDF
dec0139.hsn.de.hpc.ucar.edu 502: Error dstmbl: pft= 174 lnd_frc_mbl(p)= 6.576454038243204E+031
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: local patch index = 174
dec0253.hsn.de.hpc.ucar.edu 89: Error dstmbl: pft= 58 lnd_frc_mbl(p)= 2.753951402068762E+020
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: local patch index = 58
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: global patch index = 186684
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: global column index = 108680
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: global patch index = 51815
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: global landunit index = 64616
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: global column index = 29242
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: global landunit index = 17570
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: global gridcell index = 26087
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: gridcell longitude = 256.3250000
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: global gridcell index = 7107
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: gridcell latitude = 35.9750000
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: gridcell longitude = 253.3250000
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: pft type = 0
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: gridcell latitude = 33.8250000
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: column type = 1
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: pft type = 0
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: landunit type = 1
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: column type = 1
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: landunit type = 1
dec0139.hsn.de.hpc.ucar.edu 502: ENDRUN:
dec0085.hsn.de.hpc.ucar.edu 296: Error dstmbl: pft= 81 lnd_frc_mbl(p)= 3.774403431464960E+015
dec0139.hsn.de.hpc.ucar.edu 502: ERROR: ERROR in DUSTMod.F90 at line 365
dec0253.hsn.de.hpc.ucar.edu 89: ENDRUN:
dec0085.hsn.de.hpc.ucar.edu 296: iam = 296: local patch index = 81
dec0253.hsn.de.hpc.ucar.edu 89: ERROR: ERROR in DUSTMod.F90 at line 365
dec0085.hsn.de.hpc.ucar.edu 296: iam = 296: global patch index = 83305
dec0081.hsn.de.hpc.ucar.edu 226: Error dstmbl: pft= 5 lnd_frc_mbl(p)= 321979374166.511
dec0081.hsn.de.hpc.ucar.edu 226: iam = 226: local patch index = 5
dec0085.hsn.de.hpc.ucar.edu 296: iam = 296: global column index = 46898
dec0139.hsn.de.hpc.ucar.edu 386: Error dstmbl: pft= 5 lnd_frc_mbl(p)= 221199400734.272
dec0253.hsn.de.hpc.ucar.edu 3: Error dstmbl: pft= 63 lnd_frc_mbl(p)= 28693065948.9600
dec0081.hsn.de.hpc.ucar.edu 226: iam = 226: global patch index = 10654
Could the issue be due to the mismatch in spatial resolution between the MODIS LAI data (0.5°) and the surface dataset (0.05°)? Is it possible to regrid the MODIS LAI data to match the 0.05° resolution? Additionally, the surface dataset is configured with 78 plant functional types (PFTs), while the MODIS LAI data uses only 16 PFTs. Could this PFT mismatch also be causing the error?
My goal is to use monthly prescribed LAI that varies interannually while retaining the fine spatial resolution. Is there any way to make this configuration work? Alternatively, is there a surface dataset available that provides interannually varying LAI at high spatial resolution?
For the surface dataset, I used:
/glade/campaign/cgd/tss/people/oleson/CLM5_datasets/ctsm5.2.0/GLOBAL/surfdata_0.05x0.05-hires_hist_2005_78pfts_c240425.nc
because I want to run the model at a fine resolution. Since the monthly LAI data in the surface dataset follows the same annual cycle each year, I initially enabled the use of external LAI by setting use_lai_streams = .true. in user_nl_clm.
However, after submitting the case, I encountered an error.
/glade/derecho/scratch/koushanm/April_30_USA_MODIS_SP/run> vi cesm.log.9370208.desched1.250430-213018
dec1906.hsn.de.hpc.ucar.edu 175: # of NaNs = 1
dec1906.hsn.de.hpc.ucar.edu 175: Which are NaNs = F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F F T F F F
dec1906.hsn.de.hpc.ucar.edu 175: F F F F F F F F F F F F F F F F F F F F F F F F F F F F F
dec1906.hsn.de.hpc.ucar.edu 175: NaN found in field Sl_t at gridcell index/lon/lat: 188
dec1906.hsn.de.hpc.ucar.edu 175: 271.575000000000 43.8750000000000
dec1906.hsn.de.hpc.ucar.edu 175: ERROR: ERROR: One or more of the CTSM cap export_1D fields are NaN
dec1906.hsn.de.hpc.ucar.edu 175: Image PC Routine Line Source
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 000000000107373D shr_abort_mod_mp_ 114 shr_abort_mod.F90
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 00000000005A3E3D lnd_import_export 169 lnd_import_export_utils.F90
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 00000000005A3040 lnd_import_export 1193 lnd_import_export.F90
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 00000000005A0618 lnd_import_export 780 lnd_import_export.F90
dec1906.hsn.de.hpc.ucar.edu 175: cesm.exe 0000000000590083 lnd_comp_nuopc_mp 912 lnd_comp_nuopc.F90
In the next step, I attempted to crop the MODIS LAI stream data using the following command:
ncks -O -d lat,33,46 -d lon,250,272 /glade/u/home/koushanm/MODISPFTLAI_0.5x0.5_c140711.nc MODISPFTLAI_cropped.nc
I also used a mesh file created based on the surface dataset and added the following settings to user_nl_clm:
stream_fldfilename_lai = '/glade/u/home/koushanm/MODISPFTLAI_cropped.nc'
stream_meshfile_lai = '/glade/work/koushanm/ctsm5.2.005/CTSM/tools/site_and_regional/subset_data_regional/lnd_mesh.nc'
After submitting the case again, I encountered another error. The new log file is located at:
/glade/derecho/scratch/koushanm/April_30_USA_MODIS_SP/run/cesm.log.9370835.desched1.250430-222056
dec0253.hsn.de.hpc.ucar.edu 0: PIO2 pio_file.c retry NETCDF
dec0139.hsn.de.hpc.ucar.edu 502: Error dstmbl: pft= 174 lnd_frc_mbl(p)= 6.576454038243204E+031
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: local patch index = 174
dec0253.hsn.de.hpc.ucar.edu 89: Error dstmbl: pft= 58 lnd_frc_mbl(p)= 2.753951402068762E+020
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: local patch index = 58
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: global patch index = 186684
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: global column index = 108680
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: global patch index = 51815
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: global landunit index = 64616
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: global column index = 29242
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: global landunit index = 17570
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: global gridcell index = 26087
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: gridcell longitude = 256.3250000
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: global gridcell index = 7107
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: gridcell latitude = 35.9750000
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: gridcell longitude = 253.3250000
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: pft type = 0
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: gridcell latitude = 33.8250000
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: column type = 1
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: pft type = 0
dec0139.hsn.de.hpc.ucar.edu 502: iam = 502: landunit type = 1
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: column type = 1
dec0253.hsn.de.hpc.ucar.edu 89: iam = 89: landunit type = 1
dec0139.hsn.de.hpc.ucar.edu 502: ENDRUN:
dec0085.hsn.de.hpc.ucar.edu 296: Error dstmbl: pft= 81 lnd_frc_mbl(p)= 3.774403431464960E+015
dec0139.hsn.de.hpc.ucar.edu 502: ERROR: ERROR in DUSTMod.F90 at line 365
dec0253.hsn.de.hpc.ucar.edu 89: ENDRUN:
dec0085.hsn.de.hpc.ucar.edu 296: iam = 296: local patch index = 81
dec0253.hsn.de.hpc.ucar.edu 89: ERROR: ERROR in DUSTMod.F90 at line 365
dec0085.hsn.de.hpc.ucar.edu 296: iam = 296: global patch index = 83305
dec0081.hsn.de.hpc.ucar.edu 226: Error dstmbl: pft= 5 lnd_frc_mbl(p)= 321979374166.511
dec0081.hsn.de.hpc.ucar.edu 226: iam = 226: local patch index = 5
dec0085.hsn.de.hpc.ucar.edu 296: iam = 296: global column index = 46898
dec0139.hsn.de.hpc.ucar.edu 386: Error dstmbl: pft= 5 lnd_frc_mbl(p)= 221199400734.272
dec0253.hsn.de.hpc.ucar.edu 3: Error dstmbl: pft= 63 lnd_frc_mbl(p)= 28693065948.9600
dec0081.hsn.de.hpc.ucar.edu 226: iam = 226: global patch index = 10654
Could the issue be due to the mismatch in spatial resolution between the MODIS LAI data (0.5°) and the surface dataset (0.05°)? Is it possible to regrid the MODIS LAI data to match the 0.05° resolution? Additionally, the surface dataset is configured with 78 plant functional types (PFTs), while the MODIS LAI data uses only 16 PFTs. Could this PFT mismatch also be causing the error?
My goal is to use monthly prescribed LAI that varies interannually while retaining the fine spatial resolution. Is there any way to make this configuration work? Alternatively, is there a surface dataset available that provides interannually varying LAI at high spatial resolution?