Dear Scientists,
I want to run a regional IHISTClm50SpGs case using my own landuse_timeseries data, surface data and meterological data. With your patient answer and help, I successfully generated a 0.01 deg surfdata file (2000) using the rawdata and land use data I prepared. I also use dynamic LAI I prepared by setting use_lai_streams = .true.. I successfully ran a I2000Clm50SpGs case for spinup and got the restart file as the initial file for IHISTClm50SpGs case. I have indicated these files in user_nl_clm as below:
&clm_inparm
fsurdat='/home/user/cesm/inputdata/lnd/clm2/surfdata_map/surfdata_0.01x0.01_shanxi_16pfts_Irrig_simyr2000_c240401.nc'
flanduse_timeseries ='/home/user/cesm/inputdata/lnd/clm2/landuse.timeseries.nc'
finidat='/home/user/cesm/scratch/shanxispinup7/run/shanxispinup7.clm2.r.0081-01-01-00000.nc'
use_init_interp = .true.
hist_empty_htapes = .false.
use_lai_streams = .true.
hist_mfilt = 12, 30
hist_nhtfrq = 0, -24
hist_fincl1 = 'FPSN', 'H2OSOI ', 'QVEGT', 'QVEGE', 'QSOIL', 'SOILLIQ ', 'QFLX_EVAP_TOT', 'QRUNOFF', 'QOVER'
/
then I build and submit the case while I got many errors as below:
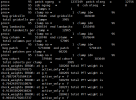
ENDRUN:
ERROR: ERROR in subgridWeightsMod.F90 at line 703
check_weights ERROR: at g = 430 total PFT weight is
0.970000000000000 active_only = F
check_weights ERROR: at g = 516 total PFT weight is
0.910000000000000 active_only = F
check_weights ERROR: at g = 517 total PFT weight is
0.820000000000000 active_only = F
check_weights ERROR: at g = 518 total PFT weight is
0.940000000000000 active_only = F
check_weights ERROR: at g = 519 total PFT weight is
0.950000000000000 active_only = F
check_weights ERROR: at g = 1130 total PFT weight is
0.980000000000000 active_only = F
check_weights ERROR: at g = 430 total col weight is
0.970000000000000 active_only = F
check_weights ERROR: at g = 516 total col weight is
0.910000000000000 active_only = F
check_weights ERROR: at g = 517 total col weight is
0.820000000000000 active_only = F
check_weights ERROR: at g = 518 total col weight is
0.940000000000000 active_only = F
check_weights ERROR: at g = 519 total col weight is
0.950000000000000 active_only = F
check_weights ERROR: at g = 1130 total col weight is
0.980000000000000 active_only = F
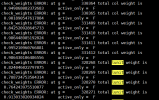
check_weights ERROR: at g = 430 total lunit weight is
0.970000000000000 active_only = F
check_weights ERROR: at g = 516 total lunit weight is
0.910000000000000 active_only = F
check_weights ERROR: at g = 517 total lunit weight is
0.820000000000000 active_only = F
check_weights ERROR: at g = 518 total lunit weight is
0.940000000000000 active_only = F
check_weights ERROR: at g = 519 total lunit weight is
0.950000000000000 active_only = F
check_weights ERROR: at g = 1130 total lunit weight is
0.980000000000000 active_only = F
If you are seeing this message at the beginning of a run with
use_init_interp = .true. and init_interp_method = "use_finidat_areas",
and you are seeing weights less than 1, then a likely cause is:
For the above-mentioned grid cell(s):
The matching input grid cell had some non-zero-weight subgrid type
that is not present in memory in the new run.
If you are using a ctsm5.2 or later fsurdat file containing
PCT_OCEAN > 0, then you may solve the error by setting
convert_ocean_to_land = .true.
As the surfdata I use in this case is absolutely the same as that in the spin up case, and the spin up case has been successfully run. I think the error should not point to the surfdata. And I also checked the pft data in the landuse_timeseries file, I think it should be fine. Do you have any suggestions for this error?
Any advice would be greatly appreciated. Thank you!
I want to run a regional IHISTClm50SpGs case using my own landuse_timeseries data, surface data and meterological data. With your patient answer and help, I successfully generated a 0.01 deg surfdata file (2000) using the rawdata and land use data I prepared. I also use dynamic LAI I prepared by setting use_lai_streams = .true.. I successfully ran a I2000Clm50SpGs case for spinup and got the restart file as the initial file for IHISTClm50SpGs case. I have indicated these files in user_nl_clm as below:
&clm_inparm
fsurdat='/home/user/cesm/inputdata/lnd/clm2/surfdata_map/surfdata_0.01x0.01_shanxi_16pfts_Irrig_simyr2000_c240401.nc'
flanduse_timeseries ='/home/user/cesm/inputdata/lnd/clm2/landuse.timeseries.nc'
finidat='/home/user/cesm/scratch/shanxispinup7/run/shanxispinup7.clm2.r.0081-01-01-00000.nc'
use_init_interp = .true.
hist_empty_htapes = .false.
use_lai_streams = .true.
hist_mfilt = 12, 30
hist_nhtfrq = 0, -24
hist_fincl1 = 'FPSN', 'H2OSOI ', 'QVEGT', 'QVEGE', 'QSOIL', 'SOILLIQ ', 'QFLX_EVAP_TOT', 'QRUNOFF', 'QOVER'
/
then I build and submit the case while I got many errors as below:
ENDRUN:
ERROR: ERROR in subgridWeightsMod.F90 at line 703
check_weights ERROR: at g = 430 total PFT weight is
0.970000000000000 active_only = F
check_weights ERROR: at g = 516 total PFT weight is
0.910000000000000 active_only = F
check_weights ERROR: at g = 517 total PFT weight is
0.820000000000000 active_only = F
check_weights ERROR: at g = 518 total PFT weight is
0.940000000000000 active_only = F
check_weights ERROR: at g = 519 total PFT weight is
0.950000000000000 active_only = F
check_weights ERROR: at g = 1130 total PFT weight is
0.980000000000000 active_only = F
check_weights ERROR: at g = 430 total col weight is
0.970000000000000 active_only = F
check_weights ERROR: at g = 516 total col weight is
0.910000000000000 active_only = F
check_weights ERROR: at g = 517 total col weight is
0.820000000000000 active_only = F
check_weights ERROR: at g = 518 total col weight is
0.940000000000000 active_only = F
check_weights ERROR: at g = 519 total col weight is
0.950000000000000 active_only = F
check_weights ERROR: at g = 1130 total col weight is
0.980000000000000 active_only = F
check_weights ERROR: at g = 430 total lunit weight is
0.970000000000000 active_only = F
check_weights ERROR: at g = 516 total lunit weight is
0.910000000000000 active_only = F
check_weights ERROR: at g = 517 total lunit weight is
0.820000000000000 active_only = F
check_weights ERROR: at g = 518 total lunit weight is
0.940000000000000 active_only = F
check_weights ERROR: at g = 519 total lunit weight is
0.950000000000000 active_only = F
check_weights ERROR: at g = 1130 total lunit weight is
0.980000000000000 active_only = F
If you are seeing this message at the beginning of a run with
use_init_interp = .true. and init_interp_method = "use_finidat_areas",
and you are seeing weights less than 1, then a likely cause is:
For the above-mentioned grid cell(s):
The matching input grid cell had some non-zero-weight subgrid type
that is not present in memory in the new run.
If you are using a ctsm5.2 or later fsurdat file containing
PCT_OCEAN > 0, then you may solve the error by setting
convert_ocean_to_land = .true.
As the surfdata I use in this case is absolutely the same as that in the spin up case, and the spin up case has been successfully run. I think the error should not point to the surfdata. And I also checked the pft data in the landuse_timeseries file, I think it should be fine. Do you have any suggestions for this error?
Any advice would be greatly appreciated. Thank you!