Dear all,
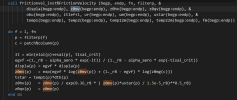
When I modified the thermodynamic roughness (z0mv) parameterization scheme, the following problems appeared:
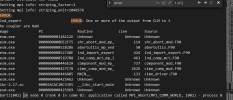
cesm.log:
ERROR:
lnd_export ERROR: One or more of the output from CLM to t
he coupler are NaN
Image PC Routine Line Source
cesm.exe 00000000012A0CC6 Unknown Unknown Unknown
cesm.exe 0000000000DE2B15 shr_abort_mod_mp_ 114 shr_abort_mod.F90
cesm.exe 0000000000528286 abortutils_mp_end 50 abortutils.F90
cesm.exe 000000000052748E lnd_import_export 424 lnd_import_export.F90
cesm.exe 000000000051D7B8 lnd_comp_mct_mp_l 463 lnd_comp_mct.F90
cesm.exe 0000000000446E2A component_mod_mp_ 737 component_mod.F90
cesm.exe 0000000000427C7B cime_comp_mod_mp_ 2626 cime_comp_mod.F90
cesm.exe 0000000000446A05 MAIN__ 133 cime_driver.F90
cesm.exe 0000000000425A62 Unknown Unknown Unknown
libc-2.17.so 00002ADB1DD163D5 __libc_start_main Unknown Unknown
cesm.exe 0000000000425969 Unknown Unknown Unknown
Abort(1001) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 1001) - process 0
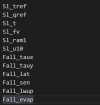
The missing values are shown in the attached picture,Hope to get your help, thank you.
When I modified the thermodynamic roughness (z0mv) parameterization scheme, the following problems appeared:
cesm.log:
ERROR:
lnd_export ERROR: One or more of the output from CLM to t
he coupler are NaN
Image PC Routine Line Source
cesm.exe 00000000012A0CC6 Unknown Unknown Unknown
cesm.exe 0000000000DE2B15 shr_abort_mod_mp_ 114 shr_abort_mod.F90
cesm.exe 0000000000528286 abortutils_mp_end 50 abortutils.F90
cesm.exe 000000000052748E lnd_import_export 424 lnd_import_export.F90
cesm.exe 000000000051D7B8 lnd_comp_mct_mp_l 463 lnd_comp_mct.F90
cesm.exe 0000000000446E2A component_mod_mp_ 737 component_mod.F90
cesm.exe 0000000000427C7B cime_comp_mod_mp_ 2626 cime_comp_mod.F90
cesm.exe 0000000000446A05 MAIN__ 133 cime_driver.F90
cesm.exe 0000000000425A62 Unknown Unknown Unknown
libc-2.17.so 00002ADB1DD163D5 __libc_start_main Unknown Unknown
cesm.exe 0000000000425969 Unknown Unknown Unknown
Abort(1001) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 1001) - process 0
The missing values are shown in the attached picture,Hope to get your help, thank you.