Dear all,
I am trying to subset a 0.125x0.125 surfdata (obtained from https://svn-ccsm-inputdata.cgd.ucar...fdata_0.125x0.125_78pfts_simyr2000_c151014.nc) using the command:
I have used this tool successfully several times, not sure why this happened. Can anyone give me a hint here? Thanks in advance.
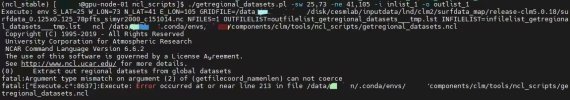
I am trying to subset a 0.125x0.125 surfdata (obtained from https://svn-ccsm-inputdata.cgd.ucar...fdata_0.125x0.125_78pfts_simyr2000_c151014.nc) using the command:
and the error occured:./getregional_datasets.pl -sw 25,73 -ne 41,105 -i inlist_1 -o outlist_1

I have used this tool successfully several times, not sure why this happened. Can anyone give me a hint here? Thanks in advance.
