Dear
@oleson thank you for your reply.
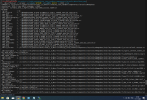
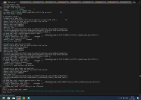
I have run the command just like you but facing the same error as shown below.
./mksurfdata.pl -r usrspec -usr_gname $GRIDNAME -usr_gdate $CDATE -y 2000 -crop
CSMDATA is /home/cas/faculty/dilipganguly/cesm/inputdata
resolution: IITD_1 rcp=-999.9 sim_year = 2000
namelist: surfdata_IITD_1_78pfts_CMIP6_simyr2000_c221107.namelist
&clmexp
nglcec = 10
mksrf_fgrid = '../mkmapdata/map_0.25x0.25_MODIS_to_IITD_1_nomask_aave_da_c221105.nc'
map_fpft = '../mkmapdata/map_0.25x0.25_MODIS_to_IITD_1_nomask_aave_da_c221105.nc'
map_fglacier = '../mkmapdata/map_3x3min_GLOBE-Gardner_to_IITD_1_nomask_aave_da_c221105.nc'
map_fglacierregion = '../mkmapdata/map_10x10min_nomask_to_IITD_1_nomask_aave_da_c221105.nc'
map_fsoicol = '../mkmapdata/map_0.25x0.25_MODIS_to_IITD_1_nomask_aave_da_c221105.nc'
map_furban = '../mkmapdata/map_3x3min_LandScan2004_to_IITD_1_nomask_aave_da_c221105.nc'
map_fmax = '../mkmapdata/map_3x3min_USGS_to_IITD_1_nomask_aave_da_c221105.nc'
map_forganic = '../mkmapdata/map_5x5min_ISRIC-WISE_to_IITD_1_nomask_aave_da_c221105.nc'
map_flai = '../mkmapdata/map_0.25x0.25_MODIS_to_IITD_1_nomask_aave_da_c221105.nc'
map_fharvest = '../mkmapdata/map_0.25x0.25_MODIS_to_IITD_1_nomask_aave_da_c221105.nc'
map_flakwat = '../mkmapdata/map_3x3min_MODIS-wCsp_to_IITD_1_nomask_aave_da_c221105.nc'
map_fwetlnd = '../mkmapdata/map_0.5x0.5_AVHRR_to_IITD_1_nomask_aave_da_c221105.nc'
map_fvocef = '../mkmapdata/map_0.5x0.5_AVHRR_to_IITD_1_nomask_aave_da_c221105.nc'
map_fsoitex = '../mkmapdata/map_5x5min_IGBP-GSDP_to_IITD_1_nomask_aave_da_c221105.nc'
map_furbtopo = '../mkmapdata/map_10x10min_nomask_to_IITD_1_nomask_aave_da_c221105.nc'
map_fgdp = '../mkmapdata/map_0.5x0.5_AVHRR_to_IITD_1_nomask_aave_da_c221105.nc'
map_fpeat = '../mkmapdata/map_0.5x0.5_AVHRR_to_IITD_1_nomask_aave_da_c221105.nc'
map_fsoildepth = '../mkmapdata/map_5x5min_ORNL-Soil_to_IITD_1_nomask_aave_da_c221105.nc'
map_fabm = '../mkmapdata/map_0.5x0.5_AVHRR_to_IITD_1_nomask_aave_da_c221105.nc'
map_ftopostats = '../mkmapdata/map_1km-merge-10min_HYDRO1K-merge-nomask_to_IITD_1_nomask_aave_da_c221105.nc'
map_fvic = '../mkmapdata/map_0.9x1.25_GRDC_to_IITD_1_nomask_aave_da_c221105.nc'
map_fch4 = '../mkmapdata/map_360x720cru_cruncep_to_IITD_1_nomask_aave_da_c221105.nc'
mksrf_fsoitex = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_soitex.10level.c010119.nc'
mksrf_forganic = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_organic_10level_5x5min_ISRIC-WISE-NCSCD_nlev7_c120830.nc'
mksrf_flakwat = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_LakePnDepth_3x3min_simyr2004_csplk_c151015.nc'
mksrf_fwetlnd = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_lanwat.050425.nc'
mksrf_fmax = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_fmax_3x3min_USGS_c120911.nc'
mksrf_fglacier = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_glacier_3x3min_simyr2000.c120926.nc'
mksrf_fglacierregion = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_GlacierRegion_10x10min_nomask_c170616.nc'
mksrf_fvocef = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_vocef_0.5x0.5_simyr2000.c110531.nc'
mksrf_furbtopo = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_topo.10min.c080912.nc'
mksrf_fgdp = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_gdp_0.5x0.5_AVHRR_simyr2000.c130228.nc'
mksrf_fpeat = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_peatf_0.5x0.5_AVHRR_simyr2000.c130228.nc'
mksrf_fsoildepth = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksf_soilthk_5x5min_ORNL-Soil_simyr1900-2015_c170630.nc'
mksrf_fabm = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_abm_0.5x0.5_AVHRR_simyr2000.c130201.nc'
mksrf_ftopostats = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_topostats_1km-merge-10min_HYDRO1K-merge-nomask_simyr2000.c130402.nc'
mksrf_fvic = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_vic_0.9x1.25_GRDC_simyr2000.c130307.nc'
mksrf_fch4 = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_ch4inversion_360x720_cruncep_simyr2000.c130322.nc'
outnc_double = .true.
all_urban = .false.
no_inlandwet = .true.
mksrf_furban = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/mksrf_urban_0.05x0.05_simyr2000.c120621.nc'
mksrf_fvegtyp = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/pftcftdynharv.0.25x0.25.LUH2.histsimyr1850-2015.c170629/mksrf_landuse_histclm50_LUH2_2000.c170629.nc'
mksrf_fhrvtyp = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/pftcftdynharv.0.25x0.25.LUH2.histsimyr1850-2015.c170629/mksrf_landuse_histclm50_LUH2_2000.c170629.nc'
mksrf_fsoicol = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/pftcftlandusedynharv.0.25x0.25.MODIS.simyr1850-2015.c170412/mksrf_soilcolor_CMIP6_simyr2005.c170623.nc'
mksrf_flai = '/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/pftcftlandusedynharv.0.25x0.25.MODIS.simyr1850-2015.c170412/mksrf_lai_78pfts_simyr2005.c170413.nc'
fsurdat = 'surfdata_IITD_1_78pfts_CMIP6_simyr2000_c221107.nc'
fsurlog = 'surfdata_IITD_1_78pfts_CMIP6_simyr2000_c221107.log'
mksrf_fdynuse = ''
fdyndat = ''
numpft = 78
/
./mksurfdata_map < surfdata_IITD_1_78pfts_CMIP6_simyr2000_c221107.namelist
Attempting to initialize control settings .....
Attempting to create surface boundary data .....
------------------------------------------------------------
mksrf_fgrid =
../mkmapdata/map_0.25x0.25_MODIS_to_IITD_1_nomask_aave_da_c221105.nc
mksrf_gridtype = global
Output ALL data in file as 64-bit
Set wetland to 0% over land
In mkpftMod::mkpftInit()...
calling domain_read
finished domain_read
fsurdat is 2d lat/lon grid
nlon= 2 nlat= 0
(OPNFIL): Successfully opened file
surfdata_IITD_1_78pfts_CMIP6_simyr2000_c221107.log on unit= 99
mksrf_gridtype = global
In mkpftMod::mkpft()...
Attempting to make PFTs .....
Creating surface datasets with extra types for crops; total pfts = 78
domain_read_dims_2d read lon and lat dims from lon/lat
domain_read initialized domain
domain_read read LANDFRAC
domain_read read LANDMASK
Open PFT file:
/home/cas/faculty/dilipganguly/cesm/inputdata/lnd/clm2/rawdata/pftcftdynharv.0.
25x0.25.LUH2.histsimyr1850-2015.c170629/mksrf_landuse_histclm50_LUH2_2000.c1706
29.nc
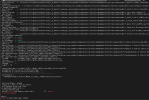
(gridmap_map_read) reading mapping matrix data...
(gridmap_map_read) * file name : ../mkmapdata/map_0.25x0.25_MODIS_to_IITD_1_nomask_aave_da_c221105.nc
* matrix dimensions rows x cols : 1036800 x 1
* number of non-zero elements: 0
(gridmap_map_read) ERROR: frac_src out of bounds
max = 1.143774191016743E+224 min = -3.094534034031665E+212
abort:
ERROR in mksurfdata_map: 34304
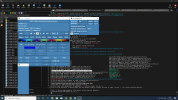
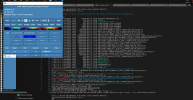
I am thinking this error is causing due to my lines in mkscripgrid.ncl script. Can you please check whether the below lines are correct or if I need to do any modifications to them?
lat(:,i) = latCenters;
latCorners(:,i,0) = latCenters - delY/2.d0;
latCorners(:,i,1) = latCenters + delY/2.d0;
latCorners(:,i,2) = latCenters + delY/2.d0;
latCorners(:,i,3) = latCenters - delY/2.d0;
end do
do j = 0, ny-1
lon(j,:) = lonCenters;
lonCorners(j,:,0) = lonCenters + delX/2.d0;
lonCorners(j,:,1) = lonCenters + delX/2.d0;
lonCorners(j,:,2) = lonCenters - delX/2.d0;
lonCorners(j,:,3) = lonCenters - delX/2.d0;
Kindly help me in this regard.