srogstad@geo_umass_edu
Member
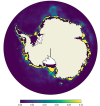
I've recently made the switch to Python after 15 years of coding in Matlab. My classes have all always been in Matlab and it is what my advisors use so I've been learning on my own through the really great NCAR Xdev tutorials and other tutorials I've found online. I have been using Xarray and Cartopy to make polar stereographic plots of CAM data and it works great, but I'm running into trouble trying to do anything with POP or CICE output. I believe it is because of the displaced pole grid, but I'm not entirely sure. When I plot CICE data on a Robinson projection it is fine but when I plot in polar stereo it is just blank. For instance here where dsice is a single monthly history output:
da = dsice['hi'].squeeze()
subplot_kws=dict(projection=ccrs.PlateCarree(),
facecolor='lightgrey')
fig=plt.figure(figsize=(16, 8))
ax1 = fig.add_subplot(1, 1, 1, projection=ccrs.SouthPolarStereo())
ax1.set_extent([-180, 180, -90, -60], crs=ccrs.PlateCarree())
p = da.plot(x='nj', y='ni',
#cmap=cm.RdBu_r, vmin=-2, vmax=2,
subplot_kws=subplot_kws,
transform=ccrs.PlateCarree())
ax1.coastlines('50m')
I tried segmenting the data (ex. p = da[:360, :].plot) after finding someone asking a similar question about plotting CICE data on another forum but that didn't work and also wouldn't help me for plotting Arctic data. When I try to plot POP data the kernel always crashes. What I'm trying is first extracting a 30 year section of the TEMP variable, taking the mean, then picking one depth slice so all I have left is a 320x384 array. I figured that would plot easy enough, but it inevitably crashes the kernel and this is the only time I've run into that issue, but it happens every time I try to plot POP data. I should also note I am using CESM1/POP2 not CESM2/MOM6 and I am using downloaded time series files on my own computer, not on Cheyenne.
I would ask this question on a place like stack exchange but I figured I'd try here since I think the answers are CESM specific more than python general questions. If anyone has advice, example plotting code, or online tutotirals specific to CICE and POP please let me know. Thanks!
da = dsice['hi'].squeeze()
subplot_kws=dict(projection=ccrs.PlateCarree(),
facecolor='lightgrey')
fig=plt.figure(figsize=(16, 8))
ax1 = fig.add_subplot(1, 1, 1, projection=ccrs.SouthPolarStereo())
ax1.set_extent([-180, 180, -90, -60], crs=ccrs.PlateCarree())
p = da.plot(x='nj', y='ni',
#cmap=cm.RdBu_r, vmin=-2, vmax=2,
subplot_kws=subplot_kws,
transform=ccrs.PlateCarree())
ax1.coastlines('50m')
I tried segmenting the data (ex. p = da[:360, :].plot) after finding someone asking a similar question about plotting CICE data on another forum but that didn't work and also wouldn't help me for plotting Arctic data. When I try to plot POP data the kernel always crashes. What I'm trying is first extracting a 30 year section of the TEMP variable, taking the mean, then picking one depth slice so all I have left is a 320x384 array. I figured that would plot easy enough, but it inevitably crashes the kernel and this is the only time I've run into that issue, but it happens every time I try to plot POP data. I should also note I am using CESM1/POP2 not CESM2/MOM6 and I am using downloaded time series files on my own computer, not on Cheyenne.
I would ask this question on a place like stack exchange but I figured I'd try here since I think the answers are CESM specific more than python general questions. If anyone has advice, example plotting code, or online tutotirals specific to CICE and POP please let me know. Thanks!