Hi,
I am trying to initialize a CESM V2.1.3 compset G1850ECOIAF run with CISO tracers in MARBL using a modified initial conditions nc file that combines ecosys_jan_IC_gx3v7_20180308.nc with 14 CISO tracers from a 9963-9982 simulated yr run that is a 20-yr average output. The CISO tracers are diat13C, diat14C, diaz13C, diaz14C, spCa13CO3, spCa14CO3, zootot13C, zootot14C, sp13C, sp14C, DO14Ctot, DO13Ctot, DI13C, and DI14C. The goal is to not have to simulate carbon isotopes out to 10,000 yrs and just simulate with a CISO 10,000 yr spun-up run. Below are my 2 runs error results with the new initial condition file:
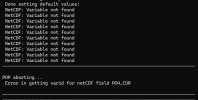
My jsl.091 model initialized, but gets DNS after a minute with the error below from cesm.log.21999.240622-110634:
Which I assume the overflow is from the 1e60.
To make the file I grabbed ecosys_jan_IC_gx3v7_20180308.nc and duplicated diatC, diazC, spCaCO3, zootot, spC, DOCtot, DIC and renamed to 13C and 14C equivalents. I copied the CISO variables from jsl.012.pop.h.9963_9983.nc into the new initial condition file called jsl012_ecosys_jan_IC_gx3v7_20180308_ciso.nc in /mnt/lustre/letscher/jsl1063/initcond/ciso9962y
I also removed time dimension and the z_t_150 depth dimension, which I converted to the z_t depth with 0 values for all the depths below depth 15-60. This dimension change is from my previous post that I did not get to till now, but refers to removing NaNs at KMT+1. Note, the error is same as my last error message from the previous post with jsl.041.
The updated model runs now have changes to DOMr_reminR_photo = 248 yr, modification to eps_dic_g equation, and iron flux modifications, which have all worked previously. I have also successfully changed the Jint_Ctot_thres within a few orders of magnitude.
An ideas on how I can fix the threshold MARBL error to initialize with this modified CISO initial conditions file?
I am trying to initialize a CESM V2.1.3 compset G1850ECOIAF run with CISO tracers in MARBL using a modified initial conditions nc file that combines ecosys_jan_IC_gx3v7_20180308.nc with 14 CISO tracers from a 9963-9982 simulated yr run that is a 20-yr average output. The CISO tracers are diat13C, diat14C, diaz13C, diaz14C, spCa13CO3, spCa14CO3, zootot13C, zootot14C, sp13C, sp14C, DO14Ctot, DO13Ctot, DI13C, and DI14C. The goal is to not have to simulate carbon isotopes out to 10,000 yrs and just simulate with a CISO 10,000 yr spun-up run. Below are my 2 runs error results with the new initial condition file:
My jsl.091 model initialized, but gets DNS after a minute with the error below from cesm.log.21999.240622-110634:
- (Task 6, block 1) MARBL ERROR (marbl_diagnostics_mod:store_diagnostics_carbon_fluxes): abs(Jint_Ctot)= 0.957E+038 exceeds Jint_Ctot_thres= 0.317E-011
- (Task 15, block 1) MARBL ERROR (marbl_ciso_diagnostics_mod:store_diagnostics_ciso_interior): abs(CISO_Jint_13Ctot)= 0.283E-003 exceeds CISO_Jint_13Ctot_thres= 0.317E-011
- Jint_Ctot_thres = 1.0e9_r8 * mpercm**2 * yps * Jint_Ctot_thres_molpm2pyr * 1e60
- CISO_Jint_14Ctot_thres = R14C_std * Jint_Ctot_thres * 1e10
Which I assume the overflow is from the 1e60.
To make the file I grabbed ecosys_jan_IC_gx3v7_20180308.nc and duplicated diatC, diazC, spCaCO3, zootot, spC, DOCtot, DIC and renamed to 13C and 14C equivalents. I copied the CISO variables from jsl.012.pop.h.9963_9983.nc into the new initial condition file called jsl012_ecosys_jan_IC_gx3v7_20180308_ciso.nc in /mnt/lustre/letscher/jsl1063/initcond/ciso9962y
I also removed time dimension and the z_t_150 depth dimension, which I converted to the z_t depth with 0 values for all the depths below depth 15-60. This dimension change is from my previous post that I did not get to till now, but refers to removing NaNs at KMT+1. Note, the error is same as my last error message from the previous post with jsl.041.
The updated model runs now have changes to DOMr_reminR_photo = 248 yr, modification to eps_dic_g equation, and iron flux modifications, which have all worked previously. I have also successfully changed the Jint_Ctot_thres within a few orders of magnitude.
An ideas on how I can fix the threshold MARBL error to initialize with this modified CISO initial conditions file?