liurj04@gmail_com
Member
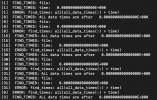
Hi, I am running CAM4 (CESM1.2.0) using prescribed bulk aerosol. When I changed the values of SO4 concentration using NCL scriptin the input aerosol data file (aero_1.9x2.5_L26_2000clim_c091112.nc) .The model can not run successfully.The error message in the log file is : READ_NEXT_TRCDATA ozone FIND_TIMES: ALL data times are after 0.000000000000000 FIND_TIMES: data times: 14.00000000000000 45.00000000000000 73.00000000000000 104.0000000000000 134.0000000000000 165.0000000000000 195.0000000000000 226.0000000000000 257.0000000000000 287.0000000000000 318.0000000000000 348.0000000000000 FIND_TIMES: time: 0.000000000000000(shr_sys_abort) ERROR: find_times: all(all_data_times(:) > time) aero_1.9x2.5_L26_2000clim_c091112.nc(shr_sys_abort) WARNING: calling shr_mpi_abort() and stopping How should I solve this error ? Please give me some suggestions.Thanks !