Hi ,
I am using CESM2.2.0 and CLM5.0, and I have recently been trying to replace the raw data for surfdata generation with high resolution raw data, to reduce the artifacts of output with high resolution grids. But even after I reference /cesm2.2.0/components/clm/tools/mksurfdata_map/READEME.developer, there are still some errors.
I take the ORGANIC data as an example, raw data in /inputdata/lnd/clm2/rawdata/mksrf_organic_10level_5x5min_ISRIC-WISE-NCSCD_nlev7_c120830.nc, gridfile in: /inputdata/lnd/clm2/mappingdata/grids/SCRIPgrid_5x5min_ISRIC-WISE_c111114.nc
Firstly, I wrote a high res organic nc file based on the variables and dimensions of mksrf_organic_10level_5x5min_ISRIC-WISE-NCSCD_nlev7_c120830.nc. Then, I ran the ./mknoocnmap.pl with my high resolution raw data grid infomation to produce a new SCRIP file, and put it in the inputdata path with the name of "SCRIPgrid_5x5min_ISRIC-WISE_c111114.nc".
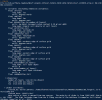
Then, I make surfdata with my destination grid,
./mknoocnmap.pl
./mkmapdata.sh
./mksurfdata.pl
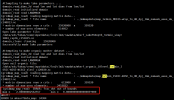
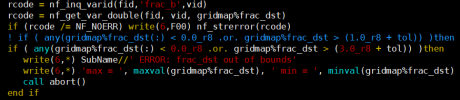
However, although the map file from grid file based on high res raw data to my destination res grid file can be generated, but it shows frac_dst is over 1 in mksurfdata process.
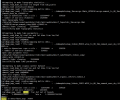
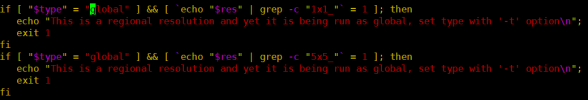
I found that the mapfile in the process of surfdata shows that the raw file is global data, but my high-resolution data is regional, so I suspect that this is the reason of errors, am I wrong?

(Note, I just replaced my nc file with the original raw data file name, then replaced the newly made SCRIP with the pathname of the original grid file, and made no changes to the code process for mkmapdata)
Appreciate for any suggestions!
I am using CESM2.2.0 and CLM5.0, and I have recently been trying to replace the raw data for surfdata generation with high resolution raw data, to reduce the artifacts of output with high resolution grids. But even after I reference /cesm2.2.0/components/clm/tools/mksurfdata_map/READEME.developer, there are still some errors.
I take the ORGANIC data as an example, raw data in /inputdata/lnd/clm2/rawdata/mksrf_organic_10level_5x5min_ISRIC-WISE-NCSCD_nlev7_c120830.nc, gridfile in: /inputdata/lnd/clm2/mappingdata/grids/SCRIPgrid_5x5min_ISRIC-WISE_c111114.nc
Firstly, I wrote a high res organic nc file based on the variables and dimensions of mksrf_organic_10level_5x5min_ISRIC-WISE-NCSCD_nlev7_c120830.nc. Then, I ran the ./mknoocnmap.pl with my high resolution raw data grid infomation to produce a new SCRIP file, and put it in the inputdata path with the name of "SCRIPgrid_5x5min_ISRIC-WISE_c111114.nc".


Then, I make surfdata with my destination grid,
./mknoocnmap.pl
./mkmapdata.sh
./mksurfdata.pl
However, although the map file from grid file based on high res raw data to my destination res grid file can be generated, but it shows frac_dst is over 1 in mksurfdata process.

I found that the mapfile in the process of surfdata shows that the raw file is global data, but my high-resolution data is regional, so I suspect that this is the reason of errors, am I wrong?

(Note, I just replaced my nc file with the original raw data file name, then replaced the newly made SCRIP with the pathname of the original grid file, and made no changes to the code process for mkmapdata)
Appreciate for any suggestions!