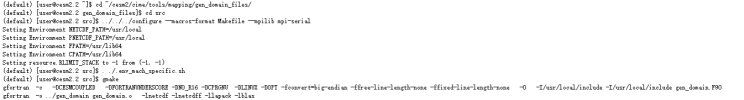Hello, everyone! I used the local atmospheric forcing dataset CMFD(China meteorological forcing dataset; 1978-2018; 0.1°×0.1°;3 hours) to replace GSWP3v1 in the CLM model (1901-2014; 0.5° x 0.5°) data. But I meet some questions.
My test was successful if I ues GSWP3v1 data, the code as follows:
My test was failed when I replace GSWP3v1 CMFD, the code as follows:

./check_input_data #no problem
./case.build
./case.submit
After that, the datm.input_data_list was deleted but Idon't know why!
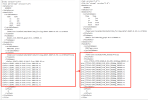
Also,./case.submit failed and the problems as follows :

I can't find the file in /home/user/cesmlab/run/SSP126_SP_QTP_0.125_04173/cesm.log.230417-142916 under the path. And I vim this file was blank.
The questions I want to ask are as follows:
Q1: Is my method of replacing data feasible, and if so, what adjustments should I make to keep my code running smoothly?
Q2: When I failed after ./case.submit, always let me see log file for details by cesm.log.XXX. But I always find it and don't know why!
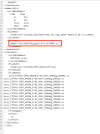
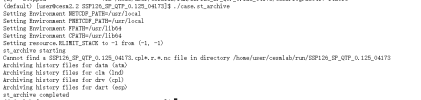
Q3. what's means of "./case.st_archive" ? the following answer appears when I run ./case.st_archive.
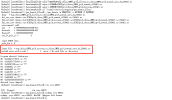
Q4. It seems like problem and how can I deal with it?
Q5: If that doesn't work, I try to write all the variables into one nc file base datm.streams.txt.CLM1PT.CLM_USRDAT. But the variables were different from my data. Is it feasible for me to modify the file of "datm. Streams. TXT. CLM1PT. CLM_USRDAT" after writing my own xxx.CLM1PT. CLM_USRDAT.nc? Will it still be replaced by the default file when I run ./case.submit
Q6:Do I need to modify any other files if I write all the variables into one nc file and changed datm. Streams. TXT. CLM1PT. CLM_USRDAT?
My test was successful if I ues GSWP3v1 data, the code as follows:
Code:
{ create_newcase --case ~/SSP126_SP_QTP_0.125_04172 --compset SSP126_DATM%GSWP3v1_CLM50%SP_SICE_SOCN_SROF_SGLC_SWAV --res CLM_USRDAT --run-unsupported
./xmlchange DIN_LOC_ROOT=/home/user/cesmlab
./xmlchange CLM_USRDAT_NAME=QTP
./xmlchange ATM_DOMAIN_FILE=domain.lnd.0.125x0.125_tx0.QTP.1v2.140704.nc,LND_DOMAIN_FILE=domain.lnd.0.125x0.125_tx0.QTP.1v2.140704.nc
./xmlchange STOP_N=1,STOP_OPTION=nyears
./xmlchange DATM_CLMNCEP_YR_START=2001,DATM_CLMNCEP_YR_END=2001
./xmlchange RUN_STARTDATE=2015-01-01
./case.setup
vim user_nl_clm
fsurdat='/home/user/cesmlab/lnd/clm2/surfdata_map/surfdata_0.125x0.125_78pfts_simyr2000_QTP_addglc_revisedLAI_c151014.nc'
flanduse_timeseries='/home/user/cesmlab/lnd/clm2/surfdata_map/release-clm5.0.18/landuse.timeseries_0.125x0.125_SSP1-2.6_78pfts_CMIP6_simyr1850-2100_QTP.nc'
finidat='/home/user/cesmlab/archive/BGC_QTP_SROF_1.1prec_3.9/rest/2015-01-01-00000/BGC_QTP_SROF_1.1prec_3.9.clm2.r.2015-01-01-00000.nc' use_init_interp = .true check_dynpft_consistency = .false hist_fincl1='TLAI'
./case.build
./case.submit}
Code:
{create_newcase --case ~/SSP126_SP_QTP_0.125_04173 --compset SSP126_DATM%GSWP3v1_CLM50%SP_SICE_SOCN_SROF_SGLC_SWAV --res CLM_USRDAT --run-unsupported
./xmlchange DIN_LOC_ROOT=/home/user/cesmlab
./xmlchange DIN_LOC_ROOT_CLMFORC=home/user/cesmlab/CMFD_modify #CMFD_modify have the files of "Precip"、"Solar"、“TPQWL”, each folder has nc files.
./xmlchange CLM_USRDAT_NAME=QTP
./xmlchange DATM_MODE="CLM1PT"
./xmlchange ATM_DOMAIN_FILE=domain.lnd.0.125x0.125_tx0.QTP.1v2.140704.nc,LND_DOMAIN_FILE=domain.lnd.0.125x0.125_tx0.QTP.1v2.140704.nc
./xmlchange STOP_N=1,STOP_OPTION=nyears
./xmlchange DATM_CLMNCEP_YR_START=2001,DATM_CLMNCEP_YR_END=2001
./xmlchange RUN_STARTDATE=2015-01-01
./case.setup
vim user_nl_clm
fsurdat='/home/user/cesmlab/lnd/clm2/surfdata_map/surfdata_0.125x0.125_78pfts_simyr2000_QTP_addglc_revisedLAI_c151014.nc'
flanduse_timeseries='/home/user/cesmlab/lnd/clm2/surfdata_map/release-clm5.0.18/landuse.timeseries_0.125x0.125_SSP1-2.6_78pfts_CMIP6_simyr1850-2100_QTP.nc'
finidat='/home/user/cesmlab/archive/BGC_QTP_SROF_1.1prec_3.9/rest/2015-01-01-00000/BGC_QTP_SROF_1.1prec_3.9.clm2.r.2015-01-01-00000.nc' use_init_interp = .true check_dynpft_consistency = .false hist_fincl1='TLAI'
./preview_namelists
#1.For the first time below the CaseDoc generated three stream file object (datm. Streams. TXT. CLM1PT. XXXX, a topo, one presaero)
#2.To model the datm. Streams. TXT. CLM1PT. 360 x720cru file as a template, create 3 stream. TXT. File:datm.streams.txt.CLM1PT.Precip; datm.streams.txt.CLM1PT.Solar; datm.streams.txt.CLM1PT.TPQWL
#3.
cp ~/SSP126_SP_QTP_0.125_04173/CaseDocs/datm.streams.txt.CLM1PT.Precip user_datm.streams.txt.CLM1PT.Precip cp ~/SSP126_SP_QTP_0.125_04173/CaseDocs/datm.streams.txt.CLM1PT.Solar user_datm.streams.txt.CLM1PT.Solar cp ~/SSP126_SP_QTP_0.125_04173/CaseDocs/datm.streams.txt.CLM1PT.TPQWL user_datm.streams.txt.CLM1PT.TPQWL
cp ~/SSP126_SP_QTP_0.125_04173/CaseDocs/datm.streams.txt.CLM1PT.Precip ~/SSP126_SP_QTP_0.125_04173/Buildconf/datmconf/datm.streams.txt.CLM1PT.Precip
cp ~/SSP126_SP_QTP_0.125_04173/CaseDocs/datm.streams.txt.CLM1PT.Solar ~/SSP126_SP_QTP_0.125_04173/Buildconf/datmconf/datm.streams.txt.CLM1PT.Solar
cp ~/SSP126_SP_QTP_0.125_04173/CaseDocs/datm.streams.txt.CLM1PT.TPQWL ~/SSP126_SP_QTP_0.125_04173/Buildconf/datmconf/datm.streams.txt.CLM1PT.TPQWL
#4.
vim user_nl_datm
&datm_nml
decomp ="1d"
factorfn = "null"
force_prognostic_true = .false.
iradsw = 1
presaero = .true.
restfilm = "undefined"
restfils = "undefined"
wiso_datm = .false.
/
&shr_strdata_nml
datamode = "CLMNCEP"
domainfile = "/home/user/cesmlab/share/domains/domain.lnd.0.125x0.125_tx0.QTP.1v2.140704.nc"
dtlimit = 1.5, 1.5, 1.5, 1.5, 1.5, 1.5 fillalgo = "nn", "nn", "nn", "nn", "nn", "nn"
fillmask = "nomask", "nomask", "nomask", "nomask", "nomask", "nomask"
fillread = "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET"
fillwrite = "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET"
mapalgo = "nn", "nn", "nn", "nn", "nn", "nn"
mapmask = "nomask", "nomask", "nomask", "nomask", "nomask", "nomask"
mapread = "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET"
mapwrite = "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET", "NOT_SET"
readmode = "single", "single", "single", "single", "single", "single"
streams = "datm.streams.txt.CLM1PT.Precip 2001 2001 2001",
"datm.streams.txt.CLM1PT.Solar 2001 2001 2001",
"datm.streams.txt.CLM1PT.TPHWL 2001 2001 2001",
"datm.streams.txt.presaero.SSP1-2.6 2015 2015 2101",
"datm.streams.txt.topo.observed 1 1 1",
"datm.streams.txt.co2tseries.SSP1-2.6 2015 2015 2500"
taxmode = "extend", "extend", "extend", "extend", "extend", "extend"
tintalgo = "nearest", "linear", "lower", "linear", "lower", "linear" vectors = "null"
/
#5.The./preview_namelists command is executed a second time, then the model updates datm_in and namelist_infile under Buildconf/datmconf according to what is in the user_nl_datm file, But the contents of the datm.input_data_list file under Buildconf/ will not be updated. so I changed the datm.input_data_list next step
./preview_namelists
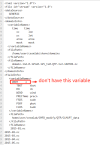
#6. changed datm.input_data_list, you can see the attach files
#7.I commented out the source code so that the contents of the datm.input_data_list file under Buildconf/ would not be overwritten
vim /opt/ncar/cesm2/cime/src/components/data_comps_mct/datm/cime_config/buildnml}
./check_input_data #no problem
./case.build
./case.submit
After that, the datm.input_data_list was deleted but Idon't know why!

Also,./case.submit failed and the problems as follows :

I can't find the file in /home/user/cesmlab/run/SSP126_SP_QTP_0.125_04173/cesm.log.230417-142916 under the path. And I vim this file was blank.
The questions I want to ask are as follows:
Q1: Is my method of replacing data feasible, and if so, what adjustments should I make to keep my code running smoothly?
Q2: When I failed after ./case.submit, always let me see log file for details by cesm.log.XXX. But I always find it and don't know why!
Q3. what's means of "./case.st_archive" ? the following answer appears when I run ./case.st_archive.
Q4. It seems like problem and how can I deal with it?

Q5: If that doesn't work, I try to write all the variables into one nc file base datm.streams.txt.CLM1PT.CLM_USRDAT. But the variables were different from my data. Is it feasible for me to modify the file of "datm. Streams. TXT. CLM1PT. CLM_USRDAT" after writing my own xxx.CLM1PT. CLM_USRDAT.nc? Will it still be replaced by the default file when I run ./case.submit
Q6:Do I need to modify any other files if I write all the variables into one nc file and changed datm. Streams. TXT. CLM1PT. CLM_USRDAT?