What I am trying to do:
I am looking at single column CAM+CLM (SCAM) and sensitivity to surface cover. I will be running the model for a year at a time. I want the crop model to be active with predictive phenology. This link shows the forcing data that I am using. The ARM data is already processed into forcing files for SCAM.
Next steps I want to do:
cesm2_3_beta17
Describe every step you took leading up to the problem:
This is part of my build script for running SCAM with a single IOP:
set CASETITLE=dyn_crop_test_case
set CASESET=FSCAM
set CASERES=T42_T42
set COMPILER=intel
set CESMROOT=/glade/work/$USER/cesm2_3_beta17
set IOP = arm97
set IOPNAME = scam_$IOP
## create new case
cd $CESMROOT/cime/scripts
#Create full casename
set CASENAME=${CASESET}.${CASERES}.${CASETITLE}
./create_newcase --case /glade/work/$USER/cases/$CASENAME --res $CASERES --compset $CASESET --compiler $COMPILER --driver nuopc --project $PROJNAME --user-mods-dir ${MODSDIR}/${IOPNAME} --run-unsupported
cd /glade/work/$USER/cases/$CASENAME
# Various XMLchange options to control settings of SCAM
./xmlchange ATM_NCPL=288 # Timesteps per day (given 5m dt)
./xmlchange STOP_OPTION=ndays
./xmlchange STOP_N=30
./xmlchange RUN_STARTDATE="2012-05-01"
./xmlchange JOB_WALLCLOCK_TIME=01:00:00
./xmlchange START_TOD=18060
./xmlchange CLM_BLDNML_OPTS="-bgc bgc -crop -ignore_ic_date -ignore_warnings"
./case.setup
I have the compset set to FSCAM, but do I need to use a specific compset to turn on the crop model?
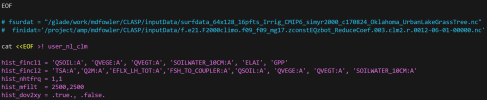
This shows the use_nl_clm from my build:
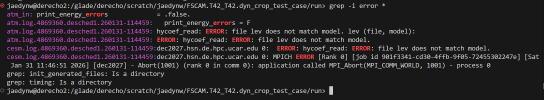
Errors that show up after submitting the case:
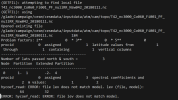
Describe your problem or question:
- Does my setup look correct in order to have the crop model running and dynamic vegetation?
- Do I need to modify assumptions about local conditions on the surface dataset and parameter files?
- Do I need to have datm files?
- It says that the file level (64) does not match the model level (32). Is there another file that you recommend I use for a test run?
I am looking at single column CAM+CLM (SCAM) and sensitivity to surface cover. I will be running the model for a year at a time. I want the crop model to be active with predictive phenology. This link shows the forcing data that I am using. The ARM data is already processed into forcing files for SCAM.
Next steps I want to do:
- Get dynamic vegetation (predictive phenology) working with the single column model to have BGC running to predict GPP, LAI, etc. From the CESM forum and wiki page, "CLM_BLDNML_OPTS" has a value of "-bgc bgc" in the env_run.xml to turn on the dynamic vegetation.
- Look at land use (monocultures versus small family-owned farms) with different crops which require having the crop model running. Are the steps in this Wiki page how I should activate the crop model? It says I can choose a compset with "crop" in the name or by adding -crop to CLM_BLDNML_OPTS.
cesm2_3_beta17
Describe every step you took leading up to the problem:
This is part of my build script for running SCAM with a single IOP:
set CASETITLE=dyn_crop_test_case
set CASESET=FSCAM
set CASERES=T42_T42
set COMPILER=intel
set CESMROOT=/glade/work/$USER/cesm2_3_beta17
set IOP = arm97
set IOPNAME = scam_$IOP
## create new case
cd $CESMROOT/cime/scripts
#Create full casename
set CASENAME=${CASESET}.${CASERES}.${CASETITLE}
./create_newcase --case /glade/work/$USER/cases/$CASENAME --res $CASERES --compset $CASESET --compiler $COMPILER --driver nuopc --project $PROJNAME --user-mods-dir ${MODSDIR}/${IOPNAME} --run-unsupported
cd /glade/work/$USER/cases/$CASENAME
# Various XMLchange options to control settings of SCAM
./xmlchange ATM_NCPL=288 # Timesteps per day (given 5m dt)
./xmlchange STOP_OPTION=ndays
./xmlchange STOP_N=30
./xmlchange RUN_STARTDATE="2012-05-01"
./xmlchange JOB_WALLCLOCK_TIME=01:00:00
./xmlchange START_TOD=18060
./xmlchange CLM_BLDNML_OPTS="-bgc bgc -crop -ignore_ic_date -ignore_warnings"
./case.setup
I have the compset set to FSCAM, but do I need to use a specific compset to turn on the crop model?
This shows the use_nl_clm from my build:

Errors that show up after submitting the case:


Describe your problem or question:
- Does my setup look correct in order to have the crop model running and dynamic vegetation?
- Do I need to modify assumptions about local conditions on the surface dataset and parameter files?
- Do I need to have datm files?
- It says that the file level (64) does not match the model level (32). Is there another file that you recommend I use for a test run?
