Good day everyone!
I have been working fairly consistently night and day the past three weeks to try to get CTSM configured for WRF on my PI's server (I'm a master's student at the University of Georgia). Via brute force and ignorance, I have reached the final stage in compiling all necessary materials for the run, that is obtaining the fsurdat file needed in the ctsm.cfg file. However, I am being blocked by a rather annoying segmentation fault, which I am failing to get around. Below is the code that I used to build the mksurfdata file:
- conda activate ctsm_pylib
- cd $HOME/WRF/CTSM/tools/mksurfdata_esmf
- export PIO=$HOME/WRF/Downloads/ParallelIO
- export PIO_INCLUDE_DIR=$PIO_PATH/include
- export PIO_LIB_DIR=$PIO_PATH/lib
- export NETCDF=$HOME/WRF/Library
- export ESMF_F90COMPILEPATHS=/opt/home/cwat/WRF/esmf/mod/modO/Linux.gfortran.64.mpiuni.default
- export MPILIB=mpich
- export FFLAGS="$FFLAGS -fallow-argument-mismatch"
- vi src/CMakeCache.txt # changed the two .so PIO files to .a. Changed ${NETCDF} to $ENV{NETCDF}.
- vi src/nanMod.F90 # fix the BOZ syntax.
- vi src/mksurfdata.F90 # I made the following changes:
#Line 271 ##added an 8 after the I [2(a,I) —> 2(a,I8)]
#LINE 289 ##added full path to pio_iotype.txt file [after compiling the first time it said it was unable to find this file. After the fix, it no longer says this.]
#Line 295 ##added an 8 after the i [(i) —> (i8)]
#Line 315 ##error that the line was too long—I split lines in two [& *returnkey* &]
#Line 328 ##add 8s after the Is in this line [(a, I, a, I) —> (a, I8, a, I8)]
- ./gen_mksurfdata_build --machine ctsm-build --verbose
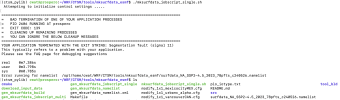
The build of this worked (the build log is attached), but I am unsure as to whether or not my little bug fixes could be contributing to the segmentation fault or not. Either way, following this I was able to successfully create the surfdata input file (attached) and make a jobscript (also attached). However, upon running ./mksurfdata_jobscript_single.sh, it gives me the following error:
(ctsm_pylib) cwat@prospero:~/WRF/CTSM/tools/mksurfdata_esmf$ ./mksurfdata_jobscript_single.sh
Attempting to initialize control settings .....
===================================================================================
= BAD TERMINATION OF ONE OF YOUR APPLICATION PROCESSES
= PID 2578958 RUNNING AT prospero
= EXIT CODE: 139
= CLEANING UP REMAINING PROCESSES
= YOU CAN IGNORE THE BELOW CLEANUP MESSAGES
===================================================================================
YOUR APPLICATION TERMINATED WITH THE EXIT STRING: Segmentation fault (signal 11)
This typically refers to a problem with your application.
Please see the FAQ page for debugging suggestions
real 0m5.461s
user 0m3.531s
sys 0m0.752s
Error running for namelist /opt/home/cwat/WRF/CTSM/tools/mksurfdata_esmf/surfdata_NA_SSP2-4.5_2023_78pfts_c240526.namelist
While I've tried to find resources to better understand this particular segmentation fault (using gdb or Valgrind), this being a BASH script accessing a file that was compiled using cmake confused Valgrind and straight-up didn't work with gdb. I've spent the past several hours trying to recompile mksurfdata while carefully tinkering around with the F90 files, but my knowledge on this stuff is very limited... I'm studying ecology, after all.
Attached I have the mksurfdata build log, a copy of the segmentation fault error (as also seen above), a copy of the jobscript, mksurfdata_esmf/src CMakeLists.txt file, and the surfdata namelist file. If anyone would like my domain file, my mesh file, and my SCRIP file, or anything else, I would be happy to send/email them. Any/all expertise would be greatly appreciated!! :)
I have been working fairly consistently night and day the past three weeks to try to get CTSM configured for WRF on my PI's server (I'm a master's student at the University of Georgia). Via brute force and ignorance, I have reached the final stage in compiling all necessary materials for the run, that is obtaining the fsurdat file needed in the ctsm.cfg file. However, I am being blocked by a rather annoying segmentation fault, which I am failing to get around. Below is the code that I used to build the mksurfdata file:
- conda activate ctsm_pylib
- cd $HOME/WRF/CTSM/tools/mksurfdata_esmf
- export PIO=$HOME/WRF/Downloads/ParallelIO
- export PIO_INCLUDE_DIR=$PIO_PATH/include
- export PIO_LIB_DIR=$PIO_PATH/lib
- export NETCDF=$HOME/WRF/Library
- export ESMF_F90COMPILEPATHS=/opt/home/cwat/WRF/esmf/mod/modO/Linux.gfortran.64.mpiuni.default
- export MPILIB=mpich
- export FFLAGS="$FFLAGS -fallow-argument-mismatch"
- vi src/CMakeCache.txt # changed the two .so PIO files to .a. Changed ${NETCDF} to $ENV{NETCDF}.
- vi src/nanMod.F90 # fix the BOZ syntax.
- vi src/mksurfdata.F90 # I made the following changes:
#Line 271 ##added an 8 after the I [2(a,I) —> 2(a,I8)]
#LINE 289 ##added full path to pio_iotype.txt file [after compiling the first time it said it was unable to find this file. After the fix, it no longer says this.]
#Line 295 ##added an 8 after the i [(i) —> (i8)]
#Line 315 ##error that the line was too long—I split lines in two [& *returnkey* &]
#Line 328 ##add 8s after the Is in this line [(a, I, a, I) —> (a, I8, a, I8)]
- ./gen_mksurfdata_build --machine ctsm-build --verbose
The build of this worked (the build log is attached), but I am unsure as to whether or not my little bug fixes could be contributing to the segmentation fault or not. Either way, following this I was able to successfully create the surfdata input file (attached) and make a jobscript (also attached). However, upon running ./mksurfdata_jobscript_single.sh, it gives me the following error:
(ctsm_pylib) cwat@prospero:~/WRF/CTSM/tools/mksurfdata_esmf$ ./mksurfdata_jobscript_single.sh
Attempting to initialize control settings .....
===================================================================================
= BAD TERMINATION OF ONE OF YOUR APPLICATION PROCESSES
= PID 2578958 RUNNING AT prospero
= EXIT CODE: 139
= CLEANING UP REMAINING PROCESSES
= YOU CAN IGNORE THE BELOW CLEANUP MESSAGES
===================================================================================
YOUR APPLICATION TERMINATED WITH THE EXIT STRING: Segmentation fault (signal 11)
This typically refers to a problem with your application.
Please see the FAQ page for debugging suggestions
real 0m5.461s
user 0m3.531s
sys 0m0.752s
Error running for namelist /opt/home/cwat/WRF/CTSM/tools/mksurfdata_esmf/surfdata_NA_SSP2-4.5_2023_78pfts_c240526.namelist
While I've tried to find resources to better understand this particular segmentation fault (using gdb or Valgrind), this being a BASH script accessing a file that was compiled using cmake confused Valgrind and straight-up didn't work with gdb. I've spent the past several hours trying to recompile mksurfdata while carefully tinkering around with the F90 files, but my knowledge on this stuff is very limited... I'm studying ecology, after all.
Attached I have the mksurfdata build log, a copy of the segmentation fault error (as also seen above), a copy of the jobscript, mksurfdata_esmf/src CMakeLists.txt file, and the surfdata namelist file. If anyone would like my domain file, my mesh file, and my SCRIP file, or anything else, I would be happy to send/email them. Any/all expertise would be greatly appreciated!! :)