I encountered the same issue, but solved it mainly according to above answers by sacks and jedwards. Share the experience incase someone need it.
First, DOWNLOADING
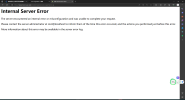
The warning occurs from the very first step to svn cesm1.2.2 to local sever.
svn co --username guestuser --password friendly https://svn-ccsm-models.cgd.ucar.edu/cesm1/release_tags/cesm1_2_2 cesm1_2_2
(1) genf90
(2) seacism_cesm
As sacks mentioned:
it can be ignored unless you're running a case with the SE dycore (e.g., ne30 resolution)
(3) mct and pio
Or
the second way is ok for downloading other github files
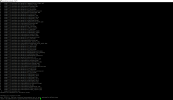
Noting that the directory of mct and pio are somewhat different. If you ignore the pio issue, it may pop up errors like:
(4) CMake
Second, Porting
As jedwards pointed out that
1.2.2.1 is not set up to handle seperate netcdf c and fortran directories so you may need to create a directory tree that links the installed libs into something that will make sence to the build, that is create netcdf/lib and netcdf/include directories, from these lnk the files from the netcdf-c and netcdf-fortran directories and set netcdf to that newly created directory. in
NETCDF issue when porting to a new machine.
Copy their lib and include to cesm_env/
Then assign the
NETCDF_PATH with
cesm_env in
cesm1_2_2/scripts/ccsm_utils/Machines/config_compilers.xml or export NETCDF_PATH to cesm_env
If NOT move netcdf-f, netcdf-fortran (and/or hdf5) together, it may shows warning/error when building:
Third, cesm_setup
Again, jedwards solved it in
error in producing namelist
you can fix the perl issues by adding a new set of parenthesis around the qw() function so:
Fourth, submitting
As alberto_sadde pointed out in
ccsm_getenv error with I_TEST_2003 in cesm1_0_4
You will need to run the script using csh your_model.run
Modify YOUR_CASE.submit to