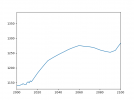
Hi all,
I am trying to calculate the change in forest area in various domains that CLM5 simulates under the various scenarios produced for CMIP6. I am using the file 'landuse.timeseries_0.9x1.25_SSP1-2.6_78pfts_CMIP6_simyr1850-2100_c190214.nc'.
Currently, I am doing this by multiplying grid cell area by PFT type and dividing by the land fraction, i.e.
AREA* PCT_NAT_PFT/100*LANDFRAC_PFT (for a given domain)
then calculating the area-weighted mean of the resulting field using CDO, adding together all the tree PFTs, and plotting. The shape of the curve looks right, but the area units don't and I am not sure why. Is this method correct or is there a better way to do this sort of thing?
Thanks,
James
I am trying to calculate the change in forest area in various domains that CLM5 simulates under the various scenarios produced for CMIP6. I am using the file 'landuse.timeseries_0.9x1.25_SSP1-2.6_78pfts_CMIP6_simyr1850-2100_c190214.nc'.
Currently, I am doing this by multiplying grid cell area by PFT type and dividing by the land fraction, i.e.
AREA* PCT_NAT_PFT/100*LANDFRAC_PFT (for a given domain)
then calculating the area-weighted mean of the resulting field using CDO, adding together all the tree PFTs, and plotting. The shape of the curve looks right, but the area units don't and I am not sure why. Is this method correct or is there a better way to do this sort of thing?
Thanks,
James