Hello,
I want to use release-clm5.0.35 complete the spin up process on my own research area, but I encountered an error in the final spin up process, here are the details of my process.
First I executed . /getregional_datasets.pl program which lodated in /gpfs/gpfs_fs/home/lmw/clm5new/tools/ncl_scripts/ to finish cropping the study area, the result is domain.lnd.fv0.9x1.25_gx1v6.090309-TP-2023-1-25.nc and surfdata_0.9x1.25_16pfts_Irrig_CMIP6_simyr1850_c170824-TP-2023-1-25.nc,
Then I created the case48 and modified the domain and surface in the lnd_in and datm_in under /gpfs/gpfs_fs/home/lmw/cesm/scratch/test48/run, so that only the study area can be spin up.
Test48 completed the 600 year AD_spin up process in the specified study area, and then I wanted to complete the final spin up in that area, according to the official documentation I copied the r file and the rpointer file
then run test53, but the error:
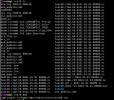
vim cesm.log.4274.230223-192214
check_dim ERROR: mismatch of input dimension 468 with expected value
20975 for variable gridcell
Did you mean to set use_init_interp = .true. in user_nl_clm?
(Setting use_init_interp = .true. is needed when doing a
transient run using an initial conditions file from a non-transient run,
or a non-transient run using an initial conditions file from a transient run,
or when running a resolution or configuration that differs from the initial conditions.)
check_dim ERROR: mismatch of input dimension 468 with expected value
20975 for variable gridcell
when I set the use_init_interp = .true. in user_nl_clm, the another error occured
vim cesm.log.4275.230223-195019
ERROR initInterp set_mindist: Cannot find any input points matching output poin
t:
subgrid level, index = pft 153031
lat, lon = -1.15959309988524 , 2.05076187109334
ltype: 6
ctype: 6
ptype: 0
Consider rerunning with the following in user_nl_clm:
init_interp_fill_missing_with_natveg = .true.
However, note that this will fill all missing types in the output
with the closest natural veg column in the input
(using bare soil for patch-level variables).
So, you should consider whether that is what you want.
maybe the reason is I can't use the r file generated by AD_spin up after specifying the study area, such as test48.clm2.r.0601-01-01-00000.nc, how can I set it to solve this problem?
Thanks in advance.
I want to use release-clm5.0.35 complete the spin up process on my own research area, but I encountered an error in the final spin up process, here are the details of my process.
First I executed . /getregional_datasets.pl program which lodated in /gpfs/gpfs_fs/home/lmw/clm5new/tools/ncl_scripts/ to finish cropping the study area, the result is domain.lnd.fv0.9x1.25_gx1v6.090309-TP-2023-1-25.nc and surfdata_0.9x1.25_16pfts_Irrig_CMIP6_simyr1850_c170824-TP-2023-1-25.nc,
Then I created the case48 and modified the domain and surface in the lnd_in and datm_in under /gpfs/gpfs_fs/home/lmw/cesm/scratch/test48/run, so that only the study area can be spin up.
./create_newcase --case test48 --res f09_g16 --compset 1850_DATM%GSWP3v1_CLM50%BGC_SICE_SOCN_SROF_SGLC_SWAV --compiler intel --mach myintel --run-unsupported
cd test 48
./xmlchange DATM_CLMNCEP_YR_START=1969
./xmlchange DATM_CLMNCEP_YR_END=1969
./xmlchange CLM_ACCELERATED_SPINUP=on
./xmlchange CLM_FORCE_COLDSTART=on
./xmlchange STOP_N=600
./xmlchange STOP_OPTION=nyears
./case.setup
./case.build
cd /gpfs/gpfs_fs/home/lmw/cesm/scratch/test48/run
vim lnd_in
fatmlndfrc = '/gpfs/gpfs_fs/home/lmw/cesm/inputdata/wodewenjian/domain.lnd.fv0.9x1.25_gx1v6.090309-TP-2023-1-25.nc'
fsurdat = '/gpfs/gpfs_fs/home/lmw/cesm/inputdata/wodewenjian/surfdata_0.9x1.25_16pfts_Irrig_CMIP6_simyr1850_c170824-TP-2023-1-25.nc'
vim datm_in
domainfile = "/gpfs/gpfs_fs/home/lmw/cesm/inputdata/wodewenjian/domain.lnd.fv0.9x1.25_gx1v6.090309-TP-2023-1-25.nc"
then run test48
Test48 completed the 600 year AD_spin up process in the specified study area, and then I wanted to complete the final spin up in that area, according to the official documentation I copied the r file and the rpointer file
./create_newcase --case test54 --res f09_g16 --compset 1850_DATM%GSWP3v1_CLM50%BGC_SICE_SOCN_SROF_SGLC_SWAV --compiler intel --mach myintel --run-unsupported
cd test54
cp /gpfs/gpfs_fs/home/lmw/cesm/scratch/test48/run/test48.clm2.r.0601-01-01-00000.nc /gpfs/gpfs_fs/home/lmw/clm5new/cime/scripts/test54
cp /gpfs/gpfs_fs/home/lmw/cesm/scratch/test48/run/rpointer.atm rpointer.drv /gpfs/gpfs_fs/home/lmw/clm5new/cime/scripts/test54
./xmlchange DATM_CLMNCEP_YR_START=1965
./xmlchange DATM_CLMNCEP_YR_END=1969
./xmlchange STOP_N=200
./xmlchange STOP_OPTION=nyears
./case.setup
echo "finidat = '/gpfs/gpfs_fs/home/lmw/clm5new/cime/scripts/test54/test48.clm2.r.0601-01-01-00000.nc' " >> ./user_nl_clm
./case.build
cd /gpfs/gpfs_fs/home/lmw/cesm/scratch/test54/run
vim lnd_in
fatmlndfrc = '/gpfs/gpfs_fs/home/lmw/cesm/inputdata/wodewenjian/domain.lnd.fv0.9x1.25_gx1v6.090309-TP-2023-1-25.nc'
fsurdat = '/gpfs/gpfs_fs/home/lmw/cesm/inputdata/wodewenjian/surfdata_0.9x1.25_16pfts_Irrig_CMIP6_simyr1850_c170824-TP-2023-1-25.nc'
vim datm_in
domainfile = "/gpfs/gpfs_fs/home/lmw/cesm/inputdata/wodewenjian/domain.lnd.fv0.9x1.25_gx1v6.090309-TP-2023-1-25.nc"
then run test53, but the error:
vim cesm.log.4274.230223-192214
check_dim ERROR: mismatch of input dimension 468 with expected value
20975 for variable gridcell
Did you mean to set use_init_interp = .true. in user_nl_clm?
(Setting use_init_interp = .true. is needed when doing a
transient run using an initial conditions file from a non-transient run,
or a non-transient run using an initial conditions file from a transient run,
or when running a resolution or configuration that differs from the initial conditions.)
check_dim ERROR: mismatch of input dimension 468 with expected value
20975 for variable gridcell
when I set the use_init_interp = .true. in user_nl_clm, the another error occured
vim cesm.log.4275.230223-195019
ERROR initInterp set_mindist: Cannot find any input points matching output poin
t:
subgrid level, index = pft 153031
lat, lon = -1.15959309988524 , 2.05076187109334
ltype: 6
ctype: 6
ptype: 0
Consider rerunning with the following in user_nl_clm:
init_interp_fill_missing_with_natveg = .true.
However, note that this will fill all missing types in the output
with the closest natural veg column in the input
(using bare soil for patch-level variables).
So, you should consider whether that is what you want.
maybe the reason is I can't use the r file generated by AD_spin up after specifying the study area, such as test48.clm2.r.0601-01-01-00000.nc, how can I set it to solve this problem?
Thanks in advance.