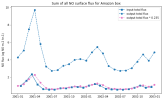
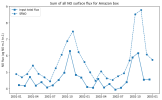
I'm running sensitivity experiments where I have increased the soil flux of NO through the input file. When I plot the emissions data from the input file, it does not match the output variable SFNO, which is supposedly the surface flux of NO.
I implemented the change by altering the namelist file:
And I see that the output surface fluxes (SFNO) respond in the right direction, but not with the magnitude that I expected. I was trying to trace through the code to figure out where SFNO is calculated, but I'm having trouble tracking that down. Could someone help me find where in the code the output SFNO field is modified relative to the input file?
CESM2
CAM6-CHEM
Land in BGC mode with fires
Prescribed SST
Historical transient simulation
compset: FCfireHIST
Thanks!
I implemented the change by altering the namelist file:
Code:
srf_emis_specifier =
(many other species listed)
'NO -> new fileAnd I see that the output surface fluxes (SFNO) respond in the right direction, but not with the magnitude that I expected. I was trying to trace through the code to figure out where SFNO is calculated, but I'm having trouble tracking that down. Could someone help me find where in the code the output SFNO field is modified relative to the input file?
CESM2
CAM6-CHEM
Land in BGC mode with fires
Prescribed SST
Historical transient simulation
compset: FCfireHIST
Thanks!