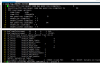
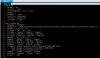
Hi, olseon, under your kind help, I have successfully replace the default GSWP3 forcing with my atm forcing (named CMFD, all of the seven forcing variable is with 3hr and 0.1° resolution) and it works. Generally, driving by the new CMFD forcing, the result is fine, however, the simulated dirunal cycle of LST (2004 average) of my study region is unreal(with a outlier in UTC 10.30 am), see attached file1. It is reasonable under the condition driviing by GSWP3, see attached file2. The CMFD data has been validated by many people. So I guess the error is attributed to my data processing or model setting. Follow your tips, I split my forcing up into three separate streams (follow the GSWP3 example) and stored in monthly format and didn't change offset, mapalgo, etc. Do your know what's wrong? Thanks a lot, you have helped me so much, I can't express how gratitude I am!
View attachment 518View attachment 519
Just curious, what CLM variable are you using to represent LST?
They don't look too different, other than the anomaly at 10:30. One thing to try is to compare the average diurnal cycle of the forcing variables (FSDS, FLDS, QBOT, TBOT, etc.) with LST and see if any of them are correlated.
Another approach is to output a set of forcing and model variables at the time resolution of the model (half-hourly or hourly). It could be that some or all of the time dimension of the forcing is not lined up with the model time. We see this sometimes when incoming solar radiation is not lined up with the cosine of the solar zenith angle in the model (i.e., when the model thinks the sun should be above or below the horizon). Although this usually shows up at the beginning or end of the model day.
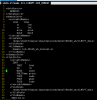
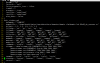
The CLM variable I am using to represent LST is 'TSKIN' . Follow your suggestion, the forcing variables (FSDS, FLDS, QBOT, TBOT) of model output variables has been output at
half-hourly time resolution(see the attached picture). I also output the
original 3hr CMFD and GSWP3
FSDS in 2004-7-31(see attached picture). I found it is the problem of
FSDS ( I used downward shortwave radiation in CMFD ). As you said, it may be caused by the reason that incoming solar radiation is not lined up with the cosine of the solar zenith angle in the model. I think its the
time-interpolation method I used in incident solar is wrong. I see in clm user guide "Example of DATM stream files with your own forcing for 3-hourly data" , the offset of prec is -5400, while solar is -10800 and others is -5400. I set it all to zero. Meanwhile, I write the time dimension of incident solar from 0 (i.e. time=0:3:669,take February as an example), do you know what's wrong and how should I do to fix it ?
Following is the matlab code to write CMFD incident solar into the format of GSWP3(take February as an example):
clc;
netcdf.setDefaultFormat('NC_FORMAT_NETCDF4')
lat0=15.05:0.1:54.95;
lon0=70.05:0.1:139.95;
for i=1:700
for j=1:400
latvalue(i,j)=lat0(j);
lonvalue(i,j)=lon0(i);
end
end
EDGEE=140;
EDGEN=55;
EDGES=15;
EDGEW=70;
LATIXY=latvalue;
LONGXY=lonvalue;
FSDS=ncread('srad_CMFD_V0106_B-01_03hr_010deg_200402.nc','srad'); %incident solar (FSDS)
FSDS=FSDS(:,:,1:224) %clm only has 28 days in February in 2004
time = (0:3:669)';
filename='solar-2004-02.nc'
ncid= netcdf.create(filename,'NC_NOCLOBBER'); %1 create nc file
%2 Defining dimensions
dimis = netcdf.defDim(ncid,'scalar',1);
dimidx = netcdf.defDim(ncid,'lon',700);
dimidy = netcdf.defDim(ncid,'lat',400);
dimit = netcdf.defDim(ncid,'time',224);
%3 Defining variables
id1 = netcdf.defVar(ncid,'EDGEE','float',[dimis]);
id2 = netcdf.defVar(ncid,'EDGEN','float',[dimis]);
id3 = netcdf.defVar(ncid,'EDGES','float',[dimis]);
id4 = netcdf.defVar(ncid,'EDGEW','float',[dimis]);
id5 = netcdf.defVar(ncid,'LATIXY','float',[dimidx dimidy]);
id6 = netcdf.defVar(ncid,'LONGXY','float',[dimidx dimidy]);
%id7 = netcdf.defVar(ncid,'TBOT','float',[dimidx dimidy dimit]);
%id8 = netcdf.defVar(ncid,'SHUM','float',[dimidx dimidy dimit]);
%id9 = netcdf.defVar(ncid,'PRECTmms','float',[dimidx dimidy dimit]);
id10 = netcdf.defVar(ncid,'FSDS','float',[dimidx dimidy dimit]);
%id11 = netcdf.defVar(ncid,'PSRF','float',[dimidx dimidy dimit]);
%id12 = netcdf.defVar(ncid,'FLDS','float',[dimidx dimidy dimit]);
%id13 = netcdf.defVar(ncid,'WIND','float',[dimidx dimidy dimit]);
%id14 = netcdf.defVar(ncid,'ZBOT','float',[dimidx dimidy dimit]);
id15 = netcdf.defVar(ncid,'time','float',[dimit]);
%4 Complete the NetCDF file definition mode
netcdf.endDef(ncid);
%5 Write the data to the NetCDF file
netcdf.putVar(ncid,id1,EDGEE)
netcdf.putVar(ncid,id2,EDGEN)
netcdf.putVar(ncid,id3,EDGES)
netcdf.putVar(ncid,id4,EDGEW)
netcdf.putVar(ncid,id5,LATIXY)
netcdf.putVar(ncid,id6,LONGXY)
%netcdf.putVar(ncid,id7,TBOT)
%netcdf.putVar(ncid,id8,SHUM)
%netcdf.putVar(ncid,id9,PRECTmms)
netcdf.putVar(ncid,id10,FSDS)
%netcdf.putVar(ncid,id11,PSRF)
%netcdf.putVar(ncid,id12,FLDS)
%netcdf.putVar(ncid,id13,WIND)
%netcdf.putVar(ncid,id14,ZBOT)
netcdf.putVar(ncid,id15,time)
netcdf.putAtt(ncid,netcdf.getConstant('NC_GLOBAL'),'title','CMFD atmospheric data at 3-hr/0.1° resolution')
netcdf.putAtt(ncid,netcdf.getConstant('NC_GLOBAL'),'conventions','CF-1.0')
netcdf.putAtt(ncid,id1,'long_name','eastern edge in atmospheric data');
netcdf.putAtt(ncid,id1,'units','degrees_east');
netcdf.putAtt(ncid,id1,'mode','time-invariant');
netcdf.putAtt(ncid,id2,'long_name','northern edge in atmospheric data');
netcdf.putAtt(ncid,id2,'units','degrees_north');
netcdf.putAtt(ncid,id2,'mode','time-invariant');
netcdf.putAtt(ncid,id3,'long_name','southern edge in atmospheric data');
netcdf.putAtt(ncid,id3,'units','degrees_north');
netcdf.putAtt(ncid,id3,'mode','time-invariant');
netcdf.putAtt(ncid,id4,'long_name','western edge in atmospheric data');
netcdf.putAtt(ncid,id4,'units','degrees_east');
netcdf.putAtt(ncid,id4,'mode','time-invariant');
netcdf.putAtt(ncid,id5,'long_name','latitude ');
netcdf.putAtt(ncid,id5,'units','degrees_north');
netcdf.putAtt(ncid,id5,'mode','time-invariant');
netcdf.putAtt(ncid,id6,'long_name','longitude ');
netcdf.putAtt(ncid,id6,'units','degrees_east');
netcdf.putAtt(ncid,id6,'mode','time-invariant');
netcdf.putAtt(ncid,id10,'long_name','incident solar (FSDS)');
netcdf.putAtt(ncid,id10,'units','W/m2');
netcdf.putAtt(ncid,id10,'mode','time-dependent');
netcdf.putAtt(ncid,id15,'long_name','observation time');
netcdf.putAtt(ncid,id15,'units','hours since 2004-02-01 00:00:00');
netcdf.putAtt(ncid,id15,'mode','time-dependent');
netcdf.putAtt(ncid,id15,'calendar','noleap');
% close file
netcdf.close(ncid);