-
The www2.cesm.ucar.edu website is currently down for maintenance
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
Lib needed for Py 2.7.15
- Thread starter wvsi3w
- Start date
git stash'git stash' will save your local changes but move them aside
Saved working directory and index state WIP on maint-5.6: 38dfe3211 update for cime5.6.44
and git pull passed too (attached).
But the "python ensemble.py --case ..." fails again:
Error: Need a valid full path with the case name (--case).
(python/3.10.13 and StdEnv/2023 are loaded, the PyCECT is updated, and the parenthesis is added to the two py files)...
Attachments
oh I see, I was using the command that I used in narval before so that's why (sorry for the confusion, I am trying 3 different systems at the same time). I created the cases directory and ran the py command, it doesn't show the previous errors, it rans a bit and stops with this new error:Does this directory exist? /home/meisam/scratch/cases/
ERROR: /lustre03/project/6001010/my_cesm_sandbox/cime/src/build_scripts/buildlib.mct FAILED, cat /home/meisam/projects/def-hbeltram/cesm2_1_3_OUT/ensemble.cesm1_tag.000/bld/mct.bldlog.231201-164715
Error building...
and when I do the cat command it shows the error as a result of not finding the netcdf paths:
gmake -f /lustre04/scratch/meisam/cases/ensemble.cesm1_tag.000/Tools/Makefile -C /home/meisam/projects/def-hbeltram/cesm2_1_3_OUT/ensemble.cesm1_tag.000/bld/intel/openmpi/nodebug/nothreads/mct CASEROOT=/lustre04/scratch/meisam/cases/ensemble.cesm1_tag.000 MODEL=mct /home/meisam/projects/def-hbeltram/cesm2_1_3_OUT/ensemble.cesm1_tag.000/bld/intel/openmpi/nodebug/nothreads/mct/Makefile.conf
gmake: Entering directory `/lustre03/project/6001010/cesm2_1_3_OUT/ensemble.cesm1_tag.000/bld/intel/openmpi/nodebug/nothreads/mct'
gmake: Leaving directory `/lustre03/project/6001010/cesm2_1_3_OUT/ensemble.cesm1_tag.000/bld/intel/openmpi/nodebug/nothreads/mct'
cat: Filepath: No such file or directory
/lustre04/scratch/meisam/cases/ensemble.cesm1_tag.000/Tools/Makefile:199: *** NETCDF not found: Define NETCDF_PATH or NETCDF_C_PATH and NETCDF_FORTRAN_PATH in config_machines.xml or config_compilers.xml. Stop.
ERROR: cat: Filepath: No such file or directory
/lustre04/scratch/meisam/cases/ensemble.cesm1_tag.000/Tools/Makefile:199: *** NETCDF not found: Define NETCDF_PATH or NETCDF_C_PATH and NETCDF_FORTRAN_PATH in config_machines.xml or config_compilers.xml. Stop.
Do you think it would be better if I use the previous loaded modules that worked in the build process, because these are Currently Loaded Modules on beluga right now which with these we have the current error:
1) CCconfig 4) gcc/12.3 (t) 7) libfabric/1.18.0 10) openmpi/4.1.5 (m) 13) StdEnv/2023 (S)
2) gentoo/2023 (S) 5) hwloc/2.9.1 8) pmix/4.2.4 11) flexiblas/3.3.1 14) mii/1.1.2
3) gcccore/.12.3 (H) 6) ucx/1.14.1 9) ucc/1.2.0 12) imkl/2023.2.0 (math) 15) python/3.10.13 (t)
and the previous (working) modules that I used in my config files are:
module load StdEnv/2018.3 perl/5.22.4 python/3.7.4 cmake/3.16.3 intelmpi/2018.3.222 hdf5-mpi/1.10.3 netcdf-mpi/4.4.1.1 netcdf-fortran-mpi/4.4.4
Hello again @jedwards ,
I have tried the ECT on both beluga and narval systems (computecanada clusters) with the modifications we discussed and the following happened:
As you remember, prior to updating PyCECT and before modifying the python codes (adding () to the codes of single_run.py and ensemble.py) the ECT didn't work on beluga. It submitted on Narval though, but failed with the error I described earlier in this thread.
After updating PyCECT and all other modifications (including using updated version of StdEnv and its modules) it showed different error on beluga (regarding the NETCDF paths not being defined in the config files, which is odd and I have defined them). And it had repeated that same error on Narval after submission. So basically only one thing changed and it was the NETCDF paths (C, FORTRAN, LIB, ...) on beluga configs.
Now, I tried it with the currently working environment on beluga and its modules (module load StdEnv/2018.3 perl/5.22.4 python/3.7.4 cmake/3.16.3 intelmpi/2018.3.222 hdf5-mpi/1.10.3 netcdf-mpi/4.4.1.1 netcdf-fortran-mpi/4.4.4) which is suitable with the configuration of the model I ran before on beluga.
However, it failed with the same error on beluga: NETCDF not found: Define NETCDF_PATH or NETCDF_C_PATH and NETCDF_FORTRAN_PATH in config_machines.xml or config_compilers.xml. Stop.
I tried it and Narval (without changing any modules or environment on Narval) and just changed the two py codes (single_run.py and ensemble.py) by adding those parenthesis to the code (like what we have on beluga) and after submitting the ECT on this system it failed after 1 hour of running with a TIMEOUT error. (last time it failed after 1 min which this means some progress!!!)
This error is new on narval and I thought maybe by increasing the walltime on this ECT I can have a complete run but by increasing the walltime nothing changed and it did run again for 1 hour and failed with timeout error again. this is odd.
one other weird thing is that last time I ran the ECT on Narval for 4 ect and all 4 were build and submitted, but this time only 1 was build and submitted.
----------------
the config files of beluga and narval are attached.
----------------
this is the ECT command on narval after I saw timeout error for the first time:
python ensemble.py --case /home/meisam/scratch/cases/ensemble2.cesm_tag.000 --mach narval --ensemble 4 --ect cam --project P99999999 --walltime 05:00:00
----------------
the "STOP_N" value for the ECT is "12", "STOP_OPTION" value is "nmonths".
First time it ran 5 months of mosart which is visible in the run directory of the case, and I thought maybe 5 hours would be enough. But by adding the --walltime option to the ECT command line it didnt change the time and stayed 1 hour !!!
----------------
Do you know why I am getting 1 ensemble run on narval instead of 4? and why it is getting timeout error when I assigned 5hours and it neglected the 5hours and ran for 1hour?
Do you think there is something wrong with the beluga config files? I asked a team from York university for this config files (beluga) and it worked on their system (Niagara cluster), and it actually worked on my system for the testing part. I tested several historical simulation using CLM5 and FATES simulation and this beluga configs worked well. I don't know if that "hard coded" paths you mentioned would be a problem here in beluga or not. I am stuck.
I have tried the ECT on both beluga and narval systems (computecanada clusters) with the modifications we discussed and the following happened:
As you remember, prior to updating PyCECT and before modifying the python codes (adding () to the codes of single_run.py and ensemble.py) the ECT didn't work on beluga. It submitted on Narval though, but failed with the error I described earlier in this thread.
After updating PyCECT and all other modifications (including using updated version of StdEnv and its modules) it showed different error on beluga (regarding the NETCDF paths not being defined in the config files, which is odd and I have defined them). And it had repeated that same error on Narval after submission. So basically only one thing changed and it was the NETCDF paths (C, FORTRAN, LIB, ...) on beluga configs.
Now, I tried it with the currently working environment on beluga and its modules (module load StdEnv/2018.3 perl/5.22.4 python/3.7.4 cmake/3.16.3 intelmpi/2018.3.222 hdf5-mpi/1.10.3 netcdf-mpi/4.4.1.1 netcdf-fortran-mpi/4.4.4) which is suitable with the configuration of the model I ran before on beluga.
However, it failed with the same error on beluga: NETCDF not found: Define NETCDF_PATH or NETCDF_C_PATH and NETCDF_FORTRAN_PATH in config_machines.xml or config_compilers.xml. Stop.
I tried it and Narval (without changing any modules or environment on Narval) and just changed the two py codes (single_run.py and ensemble.py) by adding those parenthesis to the code (like what we have on beluga) and after submitting the ECT on this system it failed after 1 hour of running with a TIMEOUT error. (last time it failed after 1 min which this means some progress!!!)
This error is new on narval and I thought maybe by increasing the walltime on this ECT I can have a complete run but by increasing the walltime nothing changed and it did run again for 1 hour and failed with timeout error again. this is odd.
one other weird thing is that last time I ran the ECT on Narval for 4 ect and all 4 were build and submitted, but this time only 1 was build and submitted.
----------------
the config files of beluga and narval are attached.
----------------
this is the ECT command on narval after I saw timeout error for the first time:
python ensemble.py --case /home/meisam/scratch/cases/ensemble2.cesm_tag.000 --mach narval --ensemble 4 --ect cam --project P99999999 --walltime 05:00:00
----------------
the "STOP_N" value for the ECT is "12", "STOP_OPTION" value is "nmonths".
First time it ran 5 months of mosart which is visible in the run directory of the case, and I thought maybe 5 hours would be enough. But by adding the --walltime option to the ECT command line it didnt change the time and stayed 1 hour !!!
----------------
Do you know why I am getting 1 ensemble run on narval instead of 4? and why it is getting timeout error when I assigned 5hours and it neglected the 5hours and ran for 1hour?
Do you think there is something wrong with the beluga config files? I asked a team from York university for this config files (beluga) and it worked on their system (Niagara cluster), and it actually worked on my system for the testing part. I tested several historical simulation using CLM5 and FATES simulation and this beluga configs worked well. I don't know if that "hard coded" paths you mentioned would be a problem here in beluga or not. I am stuck.
Attachments
Dear @jedwards
I have some good news about the ECT situation on beluga (and narval):
In the beluga setup there were some issues, mostly related to Open MPI vs Intel MPI. On the one hand I had this:
which means it'll use openmpi, but then the openmpi settings are commented out:
This is why the setup couldn't find netCDF. It is now changed this to:
<MPILIBS>intelmpi</MPILIBS>
so it'll run Intel MPI.
The Python file is modified now: by replacing "/" (division) by "//" (integer division) in ensemble.py so that it is compatible with Python 3, I don not need to load python/2.7.18 anymore.
The modules that are necessary for this configuration on my beluga system are:
module load StdEnv/2018.3 perl/5.22.4 python/3.7.4 cmake/3.16.3 intelmpi/2018.3.222 hdf5-mpi/1.10.3 netcdf-mpi/4.4.1.1 netcdf-fortran-mpi/4.4.4
Then the ECT for the 4 ensembles (python ensemble.py --case /home/meisam/scratch/cases/ensemble.cesm_tag.000 --mach beluga --ensemble 4 --ect cam --project P99999999 --walltime 12:00:00) did run and completed successfully which is very good news.
on Narval, the same Python issue occurs as on Beluga, so by doing the same // change to ensemble.py it fixed it too. Then there was a problem that the walltime isn't propagated so it uses a default walltime of 1 hour:
This could be adjusted by editing
/home/meisam/my_cesm_sandbox/cime/config/cesm/machines/config_batch.xml
and adding
Then the job successfully completed on narval system too.
+++++++++++++++++++++++++++++++++++++++++++++++++
However
+++++++++++++++++++++++++++++++++++++++++++++++++
There are some issues with the instructions regarding the ECT in your website.
For example, in this link (6. Porting and validating CIME on a new platform — CIME master documentation)
it is mentioned that you need to go through the README instructions. This is what the README says:
"Once all ensemble simulations have run successfully, copy every cam history file (*.cam.h0.*) for CAM-ECT and UF-CAM-ECT) or monthly pop history file (*.pop.h.*) for POP-ECT from each ensemble run directory into a separate directory. Next create the ensemble summary using the pyCECT tool pyEnsSum.py (for CAM-ECT and UF-CAM-ECT) or pyEnsSumPop.py (for POP-ECT). For details see README_pyEnsSum.rst and README_pyEnsSumPop.rst with the pyCECT tools."
Creating test runs: (1) Once an ensemble summary file has been created or chosen to use from $CESMDATAROOT/inputdata/validation, the simulation run(s) to be verified by ECT must be created via script ensemble.py. NOTE: It is important that the **same** resolution and compset be used in the individual runs as in the ensemble. The NetCDF ensemble summary file global attributes give this information. (2) For example, for CAM-ECT: python ensemble.py --case /glade/scratch/cesm_user/cesm_tag/camcase.cesm_tag.000 --ect cam --mach cheyenne --project P99999999 --compset F2000climo --res f19_f19_mg17 (3) Next verify the new simulation(s) with the pyCECT tool pyCECT.py (see README_pyCECT.rst with the pyCECT tools)
But in this link (Python Tools | Community Earth System Model) it says to do "./addmetadata.sh" after running the 3 runs, which contradicts the README file instructions. The 3 runs that this link states do not have ensemble numbers in them.
Also, in these 3 runs there is a typo: "./addmetadata.sh --caseroot /glade/scratch/cesm_user/cesm_tag/case.esm_tag.uf.000 --histfile /glade/scratch/cesm_user/cesm_tag/case.cesm_tag.uf.000/run/case.cesm_tag.uf.000.cam.h0.0001-01-01-00000.nc"
which I am sure instead of that esm it should be cesm.
++++++++++++++++++++++++++++++++
Then
++++++++++++++++++++++++++++++++
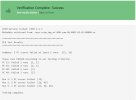
When I uploaded the three runs of ECT to the verification page it said "Verification complete-success: These runs PASSED according to our testing criterion", but there is some other vague information about some runs that failed which I would appreciate it if you check the attached JPG and let me know if it is critical fail or not.
If I am right, the ECT process and its verification is done and my porting and configuration work well (?)
Thanks for your time,
P.S. I should mention that Bart Oldeman from The Alliance Support Team helped a lot regarding these issues.
I have some good news about the ECT situation on beluga (and narval):
In the beluga setup there were some issues, mostly related to Open MPI vs Intel MPI. On the one hand I had this:
Code:
<COMPILERS>intel</COMPILERS>
<!-- MPILIBS: mpilibs supported on this machine, comma seperated list,
first is default, mpi-serial is assumed and not required in this list-->
<!-- NFT: intelmpi produces errors, so openmpi is recommended. -->
<MPILIBS>openmpi,intelmpi</MPILIBS>which means it'll use openmpi, but then the openmpi settings are commented out:
Code:
<!-- <modules mpilib="openmpi" DEBUG="FALSE">
<command name="unload">intelmpi/2018.3</command>
<command name="load">intel/2018.3</command>
<command name="load">openmpi/3.1.2</command>
<command name="load">hdf5-mpi/1.10.3</command>
<command name="load">netcdf-mpi/4.6.1</command>
<command name="load">netcdf-fortran-mpi/4.5.1</command>
</modules> -->This is why the setup couldn't find netCDF. It is now changed this to:
<MPILIBS>intelmpi</MPILIBS>
so it'll run Intel MPI.
The Python file is modified now: by replacing "/" (division) by "//" (integer division) in ensemble.py so that it is compatible with Python 3, I don not need to load python/2.7.18 anymore.
The modules that are necessary for this configuration on my beluga system are:
module load StdEnv/2018.3 perl/5.22.4 python/3.7.4 cmake/3.16.3 intelmpi/2018.3.222 hdf5-mpi/1.10.3 netcdf-mpi/4.4.1.1 netcdf-fortran-mpi/4.4.4
Then the ECT for the 4 ensembles (python ensemble.py --case /home/meisam/scratch/cases/ensemble.cesm_tag.000 --mach beluga --ensemble 4 --ect cam --project P99999999 --walltime 12:00:00) did run and completed successfully which is very good news.
on Narval, the same Python issue occurs as on Beluga, so by doing the same // change to ensemble.py it fixed it too. Then there was a problem that the walltime isn't propagated so it uses a default walltime of 1 hour:
This could be adjusted by editing
/home/meisam/my_cesm_sandbox/cime/config/cesm/machines/config_batch.xml
and adding
Code:
<submit_args>
<arg flag="--time" name="$JOB_WALLCLOCK_TIME"/>
</submit_args>Then the job successfully completed on narval system too.
+++++++++++++++++++++++++++++++++++++++++++++++++
However
+++++++++++++++++++++++++++++++++++++++++++++++++
There are some issues with the instructions regarding the ECT in your website.
For example, in this link (6. Porting and validating CIME on a new platform — CIME master documentation)
it is mentioned that you need to go through the README instructions. This is what the README says:
"Once all ensemble simulations have run successfully, copy every cam history file (*.cam.h0.*) for CAM-ECT and UF-CAM-ECT) or monthly pop history file (*.pop.h.*) for POP-ECT from each ensemble run directory into a separate directory. Next create the ensemble summary using the pyCECT tool pyEnsSum.py (for CAM-ECT and UF-CAM-ECT) or pyEnsSumPop.py (for POP-ECT). For details see README_pyEnsSum.rst and README_pyEnsSumPop.rst with the pyCECT tools."
Creating test runs: (1) Once an ensemble summary file has been created or chosen to use from $CESMDATAROOT/inputdata/validation, the simulation run(s) to be verified by ECT must be created via script ensemble.py. NOTE: It is important that the **same** resolution and compset be used in the individual runs as in the ensemble. The NetCDF ensemble summary file global attributes give this information. (2) For example, for CAM-ECT: python ensemble.py --case /glade/scratch/cesm_user/cesm_tag/camcase.cesm_tag.000 --ect cam --mach cheyenne --project P99999999 --compset F2000climo --res f19_f19_mg17 (3) Next verify the new simulation(s) with the pyCECT tool pyCECT.py (see README_pyCECT.rst with the pyCECT tools)
But in this link (Python Tools | Community Earth System Model) it says to do "./addmetadata.sh" after running the 3 runs, which contradicts the README file instructions. The 3 runs that this link states do not have ensemble numbers in them.
Also, in these 3 runs there is a typo: "./addmetadata.sh --caseroot /glade/scratch/cesm_user/cesm_tag/case.esm_tag.uf.000 --histfile /glade/scratch/cesm_user/cesm_tag/case.cesm_tag.uf.000/run/case.cesm_tag.uf.000.cam.h0.0001-01-01-00000.nc"
which I am sure instead of that esm it should be cesm.
++++++++++++++++++++++++++++++++
Then
++++++++++++++++++++++++++++++++
When I uploaded the three runs of ECT to the verification page it said "Verification complete-success: These runs PASSED according to our testing criterion", but there is some other vague information about some runs that failed which I would appreciate it if you check the attached JPG and let me know if it is critical fail or not.
If I am right, the ECT process and its verification is done and my porting and configuration work well (?)
Thanks for your time,
P.S. I should mention that Bart Oldeman from The Alliance Support Team helped a lot regarding these issues.
Attachments
Yes your ensemble did pass. The verification uses a set of quasi orthogonal model variables and a few of those variables (15,16,48,49) failed to meet the criterea for at least one of your 3 runs. This is expected. Thank you for the corrections to the Readme - If you would like to formulate them as a Pull Request it would be a great help to us.
Thanks for your response.Yes your ensemble did pass. The verification uses a set of quasi orthogonal model variables and a few of those variables (15,16,48,49) failed to meet the criterea for at least one of your 3 runs. This is expected. Thank you for the corrections to the Readme - If you would like to formulate them as a Pull Request it would be a great help to us.
Sure, but I don't know how to do that pull request, is there a link for it?