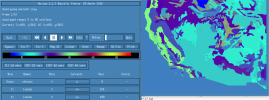
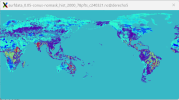
I am facing the same issue when making regional surface dataset for mid-US from ctsm5.2.mksurfdat/tools/mksurfdata_esmf. The resulting surface file has clipped data for variables PCT_CLAY, PCT_SAND, CFRAG, ORGANIC, ORGC, mapunits, etc. (see attached screenshot).
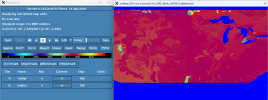
I find the same issue in different resolutions (0.05deg and 0.125deg). The surface, mesh, and logfiles are in /glade/work/amans/data/MRB-CONUS_0.125 and /glade/work/amans/data/MRB-CONUS_0.05.
Checking the logfile, I see:
Attempting to make %sand, %clay, orgc, cfrag, bulk, phaq .....
Input mapunit file is /glade/campaign/cesm/cesmdata/inputdata/lnd/clm2/rawdata/mksrf_soil_mapunits_5x5min_WISE.c220330.nc
Input lookup table file is /glade/campaign/cesm/cesmdata/inputdata/lnd/clm2/rawdata/mksrf_soil_lookup.10level.WISE.c220330.nc
Input mesh/grid file is /glade/campaign/cesm/cesmdata/inputdata/lnd/clm2/mappingdata/grids/UNSTRUCTgrid_5x5min_nomask_cdf5_c200129.nc
reading mesh_i directly in mksoiltex
Reading in mapunit data in mksoiltex
Setting mask in mesh where mapunit data is 0 mksoiltex
before route handle creation mksoiltex
after route handle creation mksoiltex
before call to dynamic mask set creation mksoiltex
after call to dynamic mask set creation mksoiltex
mksoiltex writing out mapunits
WARNING: assigning sand_o = -4 to 99%
WARNING: assigning other sand_o < 0 to 43%
WARNING: assigning clay_o = -4 to 1%
WARNING: assigning other clay_o < 0 to 18%
WARNING: assigning orgc_o = -4 to 1
WARNING: assigning other orgc_o < 0 to 0
WARNING: same warnings for cfrag_o as for orgc_o
WARNING: assigning bulk_o < 0 to 1.5
WARNING: assigning phaq_o < 0 to 7
mksoiltex writing out soil percent sand
mksoiltex writing out soil percent clay
mksoiltex writing out soil organic matter
mksoiltex writing out soil organic carbon content
mksoiltex writing out coarse fragments in soil
mksoiltex writing out soil bulk density
mksoiltex writing out soil pH measured in H2O
Successfully made %sand, %clay, orgc, cfrag, bulk, phaq .....
It looks like the issue is starting from the mapunit file. I looked at the input mapunit file but could't figure out what the issue is.