Dear all,
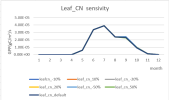
I successfully run the case I2000CLm50BgcCruGs with my own forcing data and surface data for a single-point case. However, the NEE from the outputs is higher than the flux tower data. So, I tended to modify the parameters to match the data. I changed the default s_vcad values in file "clm5_params.c200624.nc" in paramdata directory. However, the results did not change at all for the new outputs. I would like to ask why does this happen and how can I modify the parameters. THANKS!!!
I successfully run the case I2000CLm50BgcCruGs with my own forcing data and surface data for a single-point case. However, the NEE from the outputs is higher than the flux tower data. So, I tended to modify the parameters to match the data. I changed the default s_vcad values in file "clm5_params.c200624.nc" in paramdata directory. However, the results did not change at all for the new outputs. I would like to ask why does this happen and how can I modify the parameters. THANKS!!!