We generally use systems with dedicated nodes, shared node systems introduce a huge complication and frankly I just don't have any experience using them.
Hi Jim,
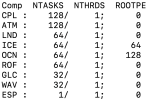
Our system administrator has installed all the prerequisites following this list:

But we got this error when testing:
xl468@dcc-hulab-01
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/scripts $ module load CESM/prereqs
OpenMPI/4.1.6
NetCDF/c-4.9.2
NetCDF/fortran-4.6.1
cmake/3.28.3
OpenBLAS 3.23
Subversion/1.14.3
CESM/prereqs
Loading
CESM/prereqs
Loading requirement: OpenMPI/4.1.6 NetCDF/c-4.9.2 NetCDF-F/fortran-4.6.1 cmake/3.28.3 OpenBLAS/3.23 Subversion/1.14.3
xl468@dcc-hulab-01
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/scripts $ module list
Currently Loaded Modulefiles:
1) OpenMPI/4.1.6 2) NetCDF/c-4.9.2 3) NetCDF-F/fortran-4.6.1 4) cmake/3.28.3 5) OpenBLAS/3.23 6) Subversion/1.14.3 7) CESM/prereqs
Key:
auto-loaded
xl468@dcc-hulab-01
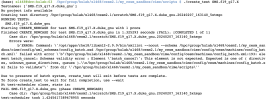
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/scripts $ ./create_test SMS.f19_g17.X
Testnames: ['SMS.f19_g17.X.duke_gnu']
No project info available
Creating test directory /hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk
RUNNING TESTS:
SMS.f19_g17.X.duke_gnu
Starting CREATE_NEWCASE for test SMS.f19_g17.X.duke_gnu with 1 procs
Finished CREATE_NEWCASE for test SMS.f19_g17.X.duke_gnu in 1.407000 seconds (PASS)
Starting XML for test SMS.f19_g17.X.duke_gnu with 1 procs
Finished XML for test SMS.f19_g17.X.duke_gnu in 0.313794 seconds (PASS)
Starting SETUP for test SMS.f19_g17.X.duke_gnu with 1 procs
Finished SETUP for test SMS.f19_g17.X.duke_gnu in 1.512333 seconds (PASS)
Starting SHAREDLIB_BUILD for test SMS.f19_g17.X.duke_gnu with 1 procs
Finished SHAREDLIB_BUILD for test SMS.f19_g17.X.duke_gnu in 4.181885 seconds (FAIL). [COMPLETED 1 of 1]
Case dir: /hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk
Errors were:
b'Building test for SMS in directory /hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk\nERROR: /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/build_scripts/buildlib.gptl FAILED, cat /hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk/bld/gptl.bldlog.240214-145317'
Due to presence of batch system, create_test will exit before tests are complete.
To force create_test to wait for full completion, use --wait
At test-scheduler close, state is:
FAIL SMS.f19_g17.X.duke_gnu (phase SHAREDLIB_BUILD)
Case dir: /hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk
test-scheduler took 7.832472324371338 seconds
xl468@dcc-hulab-01
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/scripts $ cat /hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk/bld/gptl.bldlog.240214-145317
make -f /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/Makefile install -C /hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk/bld/gnu/openmpi/nodebug/nothreads/gptl MACFILE=/hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk/Macros.make MODEL=gptl GPTL_DIR=/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing GPTL_LIBDIR=/hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk/bld/gnu/openmpi/nodebug/nothreads/gptl SHAREDPATH=/hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk/bld/gnu/openmpi/nodebug/nothreads
make: Entering directory '/hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk/bld/gnu/openmpi/nodebug/nothreads/gptl'
mpicc -c -I/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing -std=gnu99 -O -DFORTRANUNDERSCORE -DNO_R16 -DCPRGNU -DHAVE_MPI /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/gptl.c
mpicc -c -I/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing -std=gnu99 -O -DFORTRANUNDERSCORE -DNO_R16 -DCPRGNU -DHAVE_MPI /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/GPTLutil.c
mpicc -c -I/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing -std=gnu99 -O -DFORTRANUNDERSCORE -DNO_R16 -DCPRGNU -DHAVE_MPI /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/GPTLget_memusage.c
mpicc -c -I/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing -std=gnu99 -O -DFORTRANUNDERSCORE -DNO_R16 -DCPRGNU -DHAVE_MPI /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/GPTLprint_memusage.c
mpicc -c -I/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing -std=gnu99 -O -DFORTRANUNDERSCORE -DNO_R16 -DCPRGNU -DHAVE_MPI /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/gptl_papi.c
mpicc -c -I/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing -std=gnu99 -O -DFORTRANUNDERSCORE -DNO_R16 -DCPRGNU -DHAVE_MPI /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/f_wrappers.c
mpifort -c -I/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing -fconvert=big-endian -ffree-line-length-none -ffixed-line-length-none -O -DFORTRANUNDERSCORE -DNO_R16 -DCPRGNU -DHAVE_MPI -ffree-form /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/perf_utils.F90
make: Leaving directory '/hpc/group/hulab/xl468/cesm2.1/scratch/SMS.f19_g17.X.duke_gnu.20240214_145312_ldx4wk/bld/gnu/openmpi/nodebug/nothreads/gptl'
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/gptl.c: In function ‘GPTLpr_summary_file’:
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/gptl.c:3090:8: warning: cast from pointer to integer of different size [-Wpointer-to-int-cast]
3090 | if (((int) comm) == 0)
| ^
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/perf_utils.F90:282:18:
282 | call MPI_BCAST(vec,lsize,MPI_INTEGER,0,comm,ierr)
| 1
......
314 | call MPI_BCAST(vec,lsize,MPI_LOGICAL,0,comm,ierr)
| 2
Error: Type mismatch between actual argument at (1) and actual argument at (2) (INTEGER(4)/LOGICAL(4)).
make: *** [/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/Makefile:63: perf_utils.o] Error 1
ERROR: /hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/gptl.c: In function GPTLpr_summary_file :
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/gptl.c:3090:8: warning: cast from pointer to integer of different size [-Wpointer-to-int-cast]
3090 | if (((int) comm) == 0)
| ^
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/perf_utils.F90:282:18:
282 | call MPI_BCAST(vec,lsize,MPI_INTEGER,0,comm,ierr)
| 1
......
314 | call MPI_BCAST(vec,lsize,MPI_LOGICAL,0,comm,ierr)
| 2
Error: Type mismatch between actual argument at (1) and actual argument at (2) (INTEGER(4)/LOGICAL(4)).
make: *** [/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/src/share/timing/Makefile:63: perf_utils.o] Error 1xl468@dcc-hulab-01
/hpc/group/hulab/xl468/cesm2.1/my_cesm_sandbox/cime/scripts $
Here is our config_compilers.xml:

Any suggestions would be appreciated!
Thanks,
Xiang