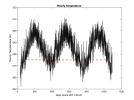
I am running CLM for a single point using atmospheric forcings from an experimental site (Xiaoji, China). The simulation completed successfully. However, most of the output variables (e.g., GDD0, GDD0, GDDPLANT) have values of 0. I also checked the temperature field (TSA, TA, A5TMIN), and these have a value of approximately 265 Kelvin which does not match the input climate forcing. The run is a startup using initial conditions taken from a global run extracted using the use_init_interp function in user_nl_clm. The plot below is the holy temperature from the input atmospheric forcing.
The details my case goes here:

The case is here /glade/work/jyotis/point_runs/i.e20.I2000Clm50BgcCrop.1pt.AgMIPOzone-xiaoji.007
The details my case goes here:
env_run.xml
Code:
./xmlchange CLM_USRDAT_NAME=1x1pt_CH-XIA
./xmlchange DATM_CLMNCEP_YR_START=2011
./xmlchange DATM_CLMNCEP_YR_END=2013
./xmlchange DATM_CLMNCEP_YR_ALIGN=1
./xmlchange MPILIB=mpi-serial
./xmlchange ATM_DOMAIN_PATH=/glade/work/jyotis/inputdata/1x1pt_CH-XIA
./xmlchange LND_DOMAIN_PATH=/glade/work/jyotis/inputdata/1x1pt_CH-XIA
./xmlchange ATM_DOMAIN_FILE=domain.lnd.1x1pt_CH-XIA_navy.220314.nc
./xmlchange LND_DOMAIN_FILE=domain.lnd.1x1pt_CH-XIA_navy.220314.nc
./xmlchange DIN_LOC_ROOT=/glade/work/jyotis/inputdata
./xmlchange DIN_LOC_ROOT_CLMFORC=/glade/work/jyotis/inputdata/atm/datm7
./xmlchange RUN_STARTDATE=2011-09-01
./xmlchange --file env_run.xml --id STOP_OPTION --val date
./xmlchange STOP_DATE=20131230
./xmlchange --file env_run.xml --id DATM_MODE --val CLM1PT
./xmlchange CLM_FORCE_COLDSTART=offuser_nl_clm
Code:
paramfile = '/glade/work/jyotis/inputdata/editparam/clm5_params.c171117.nc'
fsurdat = '/glade/work/jyotis/inputdata/lnd/clm2/surfdata_map/surfdata_1x1pt_CH-XIA_hist_78pfts_CMIP6_simyr2000_c220314.nc'
finidat = '/glade/scratch/jyotis/i.e20.I2000Clm50BgcCrop.1pt.AgMIPOzone-init-clm2.001/run/i.e20.I2000Clm50BgcCrop.1pt.AgMIPOzone-init-clm2.001.clm2.r.2011-09-01-00000.nc'
use_init_interp = .true.
hist_mfilt = 1,365,1460,8760
hist_nhtfrq = 0,-24,-6,-1
hist_fincl2 = 'FCTR','FCEV','FGEV','FSH_V','FSH_G','HBOT','HTOP','ELAI','NDEP_TO_SMINN','NFIX_TO_SMINN','SMINN_TO_PLANT','SUPPLEMENT_TO_SMINN','SMINN','C_ALLOMETRY','N_ALLOMETRY','AVAILC','PLANT_NDEMAND','PLANT_NALLOC','PLANT_CALLOC','SMINN_TO_NPOOL','RETRANSN_TO_NPOOL','TOTPFTN','PFT_NTRUNC','TOTVEGN','TLAI','TSAI','TSA','LEAFC','LEAFN','FROOTC','FROOTN','NPP','NEE','SEEDC','SEEDN','GPP','LIVESTEMC','LIVESTEMN','TOTCOLC','TOTCOLN','TOTECOSYSC','TOTECOSYSN','TOTLITC','TOTLITN','TOTPFTC','TOTPFTN','TOTSOMC','TOTSOMN','TOTVEGC','TOTVEGN','TV','EFLX_LH_TOT','A5TMIN','A10TMIN','DOWNREG','EXCESS_CFLUX','GRAINC_TO_FOOD','GRAINC','GRAINN','GDDTSOI','GDDPLANT','GDDHARV','GDD020','GDD0','GDD10','GDD1020','GDD8','GDD820','CPHASE'
hist_fincl3 = 'GDDPLANT','CPHASE','GDDHARV','GRAINC','GDDTSOI','GDD10','GDD0','GDD8','GDD020','A5TMIN','A10TMIN'
hist_fincl4 = 'GDDPLANT','CPHASE','GDDHARV','GRAINC','GDDTSOI','GDD10','GDD0','GDD8','GDD020','A5TMIN','A10TMIN'
hist_dov2xy = .true.,.false.,.true.,.false.user_datm.streams.txt.CLM1PT.CLM_USRDAT
Code:
<?xml version="1.0"?>
<file id="stream" version="1.0">
<dataSource>
GENERIC
</dataSource>
<domainInfo>
<variableNames>
time time
xc lon
yc lat
area area
mask mask
</variableNames>
<filePath>
/glade/work/jyotis/inputdata/1x1pt_CH-XIA
</filePath>
<fileNames>
domain.lnd.1x1pt_CH-XIA_navy.220314.nc
</fileNames>
</domainInfo>
<fieldInfo>
<variableNames>
ZBOT z
TBOT tbot
RH rh
WIND wind
PRECTmms precn
FSDS swdn
PSRF pbot
FLDS lwdn
</variableNames>
<filePath>
/glade/work/jyotis/inputdata/atm/datm7/CLM1PT_data/1x1pt_CH-XIA
</filePath>
<fileNames>
2011-02.nc
2011-03.nc
2011-04.nc
2011-05.nc
2011-06.nc
2011-07.nc
2011-08.nc
2011-09.nc
2011-10.nc
2011-11.nc
2011-12.nc
2012-01.nc
2012-02.nc
2012-03.nc
2012-04.nc
2012-05.nc
2012-06.nc
2012-07.nc
2012-08.nc
2012-09.nc
2012-10.nc
2012-11.nc
2012-12.nc
2013-01.nc
2013-02.nc
2013-03.nc
2013-04.nc
2013-05.nc
2013-06.nc
2013-07.nc
2013-08.nc
2013-09.nc
2013-10.nc
2013-11.nc
2013-12.nc
</fileNames>
<offset>
0
</offset>
</fieldInfo>
</file>user_nl_datm
Code:
!------------------------------------------------------------------------
! Users should ONLY USE user_nl_datm to change namelists variables
! Users should add all user specific namelist changes below in the form of
! namelist_var = new_namelist_value
! Note that any namelist variable from shr_strdata_nml and datm_nml can
! be modified below using the above syntax
! User preview_namelists to view (not modify) the output namelist in the
! directory $CASEROOT/CaseDocs
! To modify the contents of a stream txt file, first use preview_namelists
! to obtain the contents of the stream txt files in CaseDocs, and then
! place a copy of the modified stream txt file in $CASEROOT with the string
! user_ prepended.
!------------------------------------------------------------------------
&datm_nml
decomp = "1d"
factorfn = "null"
force_prognostic_true = .false.
iradsw = 1
presaero = .true.
restfilm = "undefined"
restfils = "undefined"
wiso_datm = .false.
/
&shr_strdata_nml
datamode = "CLMNCEP"
domainfile = "/glade/work/jyotis/inputdata/1x1pt_CH-XIA/domain.lnd.1x1pt_CH-XIA_navy.220314.nc"
dtlimit = 1.5, 1.5, 1.5
fillalgo = "nn", "nn", "nn"
fillmask = "nomask", "nomask", "nomask"
fillread = "NOT_SET", "NOT_SET", "NOT_SET"
fillwrite = "NOT_SET", "NOT_SET", "NOT_SET"
mapalgo = "nn", "nn", "nn"
mapmask = "nomask", "nomask", "nomask"
mapread = "NOT_SET", "NOT_SET", "NOT_SET"
mapwrite = "NOT_SET", "NOT_SET", "NOT_SET"
readmode = "single", "single", "single"
streams = "datm.streams.txt.CLM1PT.CLM_USRDAT 1 2011 2013",
"datm.streams.txt.presaero.clim_2000 1 2000 2000",
"datm.streams.txt.topo.observed 1 1 1"
taxmode = "extend", "extend", "extend"
tintalgo = "nearest", "linear", "lower"
vectors = "null"
/