Hi all,
Can anyone help with this. I am stuck to this step and feel so upset. THANKS!!!
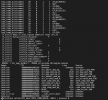
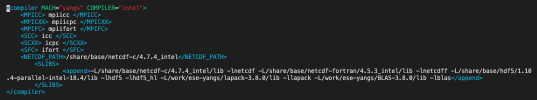
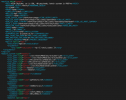
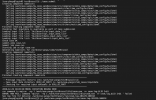
I am trying to test clm with 1x1_brazil dataset. The case was setup; build successfully. But once I did case.submit, the error showed up like below (atm.log):
dep_WACCM.ensmean_monthly_hist_1849-2015_0.9x1.25_CMIP6_c180926.nc
(shr_strdata_init_streams) timeName = 4 time
(shr_strdata_init_streams) lonName = 4 lon
(shr_strdata_init_streams) latName = 4 lat
(shr_strdata_init_streams) hgtName = 4 unknownname
(shr_strdata_init_streams) maskName = 4 mask
(shr_strdata_init_streams) areaName = 4 area
(shr_strdata_init_streams) stream 5
(shr_strdata_init_streams) filePath = 5
/work/ese-yangs/clm5_cases/inputdata/atm/datm7/topo_forcing/
(shr_strdata_init_streams) fileName = 5
/work/ese-yangs/clm5_cases/inputdata/atm/datm7/topo_forcing/topodata_0.9x1.25_U
SGS_070110_stream_c151201.nc
(shr_strdata_init_streams) timeName = 5 time
(shr_strdata_init_streams) lonName = 5 LONGXY
(shr_strdata_init_streams) latName = 5 LATIXY
(shr_strdata_init_streams) hgtName = 5 unknownname
(shr_strdata_init_streams) maskName = 5 mask
(shr_strdata_init_streams) areaName = 5 area
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for fill
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for remap
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for fill
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for remap
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for fill
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for remap
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for fill
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for remap
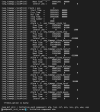
('shr_map_getWts') ERROR: yd outside bounds -90.0000000000000
('shr_map_getWts') cpole = 1.57079632679490
ERROR: ('shr_map_getWts') ERROR yd outside 90 degree bounds
Can anyone help with this. I am stuck to this step and feel so upset. THANKS!!!
I am trying to test clm with 1x1_brazil dataset. The case was setup; build successfully. But once I did case.submit, the error showed up like below (atm.log):
dep_WACCM.ensmean_monthly_hist_1849-2015_0.9x1.25_CMIP6_c180926.nc
(shr_strdata_init_streams) timeName = 4 time
(shr_strdata_init_streams) lonName = 4 lon
(shr_strdata_init_streams) latName = 4 lat
(shr_strdata_init_streams) hgtName = 4 unknownname
(shr_strdata_init_streams) maskName = 4 mask
(shr_strdata_init_streams) areaName = 4 area
(shr_strdata_init_streams) stream 5
(shr_strdata_init_streams) filePath = 5
/work/ese-yangs/clm5_cases/inputdata/atm/datm7/topo_forcing/
(shr_strdata_init_streams) fileName = 5
/work/ese-yangs/clm5_cases/inputdata/atm/datm7/topo_forcing/topodata_0.9x1.25_U
SGS_070110_stream_c151201.nc
(shr_strdata_init_streams) timeName = 5 time
(shr_strdata_init_streams) lonName = 5 LONGXY
(shr_strdata_init_streams) latName = 5 LATIXY
(shr_strdata_init_streams) hgtName = 5 unknownname
(shr_strdata_init_streams) maskName = 5 mask
(shr_strdata_init_streams) areaName = 5 area
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for fill
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for remap
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for fill
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for remap
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for fill
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for remap
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for fill
(shr_strdata_init_mapping) calling shr_dmodel_mapSet for remap
('shr_map_getWts') ERROR: yd outside bounds -90.0000000000000
('shr_map_getWts') cpole = 1.57079632679490
ERROR: ('shr_map_getWts') ERROR yd outside 90 degree bounds