Dear all,
Firstly, I successfully run the case I2000CLm50BgcCruGs with the default GSWP3v1 forcing data and the surface data which was created by CLM tools with default grids and maps. However, the output files have very low resolution for some variables (e.g., TOTECOSYSC, NEE, GPP, see attached Fig1 below). It looks like some big pixels overlap the smaller pixels in the region. Then, I used my own forcing data (0.05x0.05 degree) and I substitute the variables (PCT_PFT, PCT_CROP, PCT_NAT_PFT...) with my own higher resolution data in surface map. However, the big pixels problem is still there (see Fig2). I totally have no idea what caused these "big pixels".
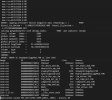
Secondly, since the above case did not run the spin-up, I tried the spin-up for the case. I used the same compset (I2000CLm50BgcCruGs) and set CLM_BLDNML_OPTS with "-bgc bgc -crop" and CLM_ACCELERATED_SPINUP="on", simulating 100 years from 2000. However, when the model run to the fifth year, it shuts down with the error: "capping procedure failed (negative mass remaining)" (see log file).
To sum up, I have two questions here. 1)why the results of TOTECOSYSC, NEE, GPP have such big pixels and how to fix it?. 2)why the spin-up shuts down and how to fix it? I am stuck to this step and I really appreciate it if anyone can help me with this. THANKS!!!
Firstly, I successfully run the case I2000CLm50BgcCruGs with the default GSWP3v1 forcing data and the surface data which was created by CLM tools with default grids and maps. However, the output files have very low resolution for some variables (e.g., TOTECOSYSC, NEE, GPP, see attached Fig1 below). It looks like some big pixels overlap the smaller pixels in the region. Then, I used my own forcing data (0.05x0.05 degree) and I substitute the variables (PCT_PFT, PCT_CROP, PCT_NAT_PFT...) with my own higher resolution data in surface map. However, the big pixels problem is still there (see Fig2). I totally have no idea what caused these "big pixels".
Secondly, since the above case did not run the spin-up, I tried the spin-up for the case. I used the same compset (I2000CLm50BgcCruGs) and set CLM_BLDNML_OPTS with "-bgc bgc -crop" and CLM_ACCELERATED_SPINUP="on", simulating 100 years from 2000. However, when the model run to the fifth year, it shuts down with the error: "capping procedure failed (negative mass remaining)" (see log file).
To sum up, I have two questions here. 1)why the results of TOTECOSYSC, NEE, GPP have such big pixels and how to fix it?. 2)why the spin-up shuts down and how to fix it? I am stuck to this step and I really appreciate it if anyone can help me with this. THANKS!!!