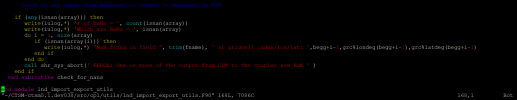
Dear Scientists,
I was running a I2000Clm50SpGs case for spinning up, but after submitting the case, an error occured and when I cat the cesm log file it shows:
# of NaNs = 7
Which are NaNs = F F F F F F T F F F F F F F F F F F F F F F F F F F F F F F F
F F F F F T T T T F F F F F T T F F F F F F F F F F F F F F
NaN found in field at gridcell index
169501
NaN found in field at gridcell index
169531
NaN found in field at gridcell index
169532
NaN found in field at gridcell index
169533
NaN found in field at gridcell index
169534
NaN found in field at gridcell index
169540
NaN found in field at gridcell index
169541
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
I wonder the reason for this error, there is no mention in the logfile why the NaN value occurred. Is this due to my atm forcing data or surfdata that contain the NaN value? Or due to my modificiation on the surfdata file? Note that I have changed the land use, PFT, LAI and soil texture data in the surfdata file but used the default SAI data. Maybe the LAI and SAI data is not corresponding in some gridcell so the NaN value occurred?
I attached the cesm.log file behind. Could you please give me some advice? Any advice would be greatly appreciated. Thank you.
I was running a I2000Clm50SpGs case for spinning up, but after submitting the case, an error occured and when I cat the cesm log file it shows:
# of NaNs = 7
Which are NaNs = F F F F F F T F F F F F F F F F F F F F F F F F F F F F F F F
F F F F F T T T T F F F F F T T F F F F F F F F F F F F F F
NaN found in field at gridcell index
169501
NaN found in field at gridcell index
169531
NaN found in field at gridcell index
169532
NaN found in field at gridcell index
169533
NaN found in field at gridcell index
169534
NaN found in field at gridcell index
169540
NaN found in field at gridcell index
169541
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
ERROR: ERROR: One or more of the output from CLM to the coupler are NaN
I wonder the reason for this error, there is no mention in the logfile why the NaN value occurred. Is this due to my atm forcing data or surfdata that contain the NaN value? Or due to my modificiation on the surfdata file? Note that I have changed the land use, PFT, LAI and soil texture data in the surfdata file but used the default SAI data. Maybe the LAI and SAI data is not corresponding in some gridcell so the NaN value occurred?
I attached the cesm.log file behind. Could you please give me some advice? Any advice would be greatly appreciated. Thank you.