Hi, everyone!
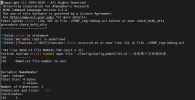
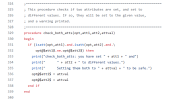
I try to test the nudging tools in directory 'components/cam/tools/nudging/Gen_Data/'.
And I chose ERAI as the target reanalysis product.
I edited the csh script according to the instructions in the README file and submitted it for execution.
Unfortunately, there was an error.
I have attached the edited csh script and the Slurm log file and hope this can provide some help.
Thanks!
I try to test the nudging tools in directory 'components/cam/tools/nudging/Gen_Data/'.
And I chose ERAI as the target reanalysis product.
I edited the csh script according to the instructions in the README file and submitted it for execution.
Unfortunately, there was an error.
I have attached the edited csh script and the Slurm log file and hope this can provide some help.
Thanks!