Hello everyone,
I am trying to run CESM2.1.3 at the resolution f05_f05_mg17. It seems there is no topo file corresponding to 0.47x0.63 available for CAM6 in https://svn-ccsm-inputdata.cgd.ucar.edu/trunk/inputdata/atm/cam/topo. I realize that the resampling tool in interpic_new can create my own topo file, but I don't find the template file for the resolution 0.47x0.63.
According to "README", I attempt to generate the template file for 0.47x0.63 resolution.
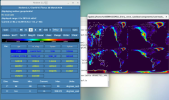
Where is the test case file baroclinic1.nc? Where can I find the template file corresponding to 0.47x0.63? Can someone help me with this problem?
Thanks in advance!
I am trying to run CESM2.1.3 at the resolution f05_f05_mg17. It seems there is no topo file corresponding to 0.47x0.63 available for CAM6 in https://svn-ccsm-inputdata.cgd.ucar.edu/trunk/inputdata/atm/cam/topo. I realize that the resampling tool in interpic_new can create my own topo file, but I don't find the template file for the resolution 0.47x0.63.
According to "README", I attempt to generate the template file for 0.47x0.63 resolution.
Bash:
% ncks -v lat,lon baroclinic1.nc SE_temp.ncThanks in advance!