I am running an ensemble spin-up using the ./CLM5_startup_freerun script included in the DART distribution on Cheyenne. I have subset my surface data files to a region in East Africa using the ./subset_surfdata python tool located in CTSM/tools/contrib for, updating the following attributes:
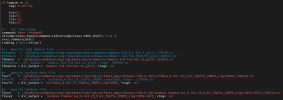
In the CLM5_startup_freerun script, I updated the setup to include the regionally subset surface data and domain files by updating the caseroot, user_nl_datm, and user_nl_clm using the following commands:
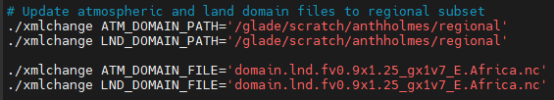


In the CLM5_startup_freerun script, I updated the setup to include the regionally subset surface data and domain files by updating the caseroot, user_nl_datm, and user_nl_clm using the following commands:


