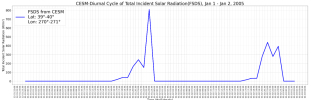
It looks like you have issues with your time intervals. For example,
ncdump -h /glade/derecho/scratch/jteymoori/Daymet4/Data/atm/datm7/Solar/clmforc.Daymet4.0.05x0.05.Solar.2005-01.nc shows there are 248 time steps in your file.
ncdump -h /glade/derecho/scratch/jteymoori/Daymet4/Data/atm/datm7/Solar/clmforc.Daymet4.0.05x0.05.Solar.2005-0w.nc ALSO shows there are 248 time steps in your file.
Thus, one issue is that Jan and Feb don't have the same number of days, so their datm files shouldn't have the same number of time steps
The second issue (that's actually seems to be causing the issue that that your time data starts on Jan 15, 2005.
Both of these issues need to be fixed in your datm files
----
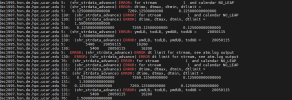
I just opened your dataset in xarray, and saw this:
ds = xr.open_dataset('/glade/derecho/scratch/jteymoori/Daymet4/Data/atm/datm7/Solar/clmforc.Daymet4.0.05x0.05.Solar.2005-01.nc',
decode_times=True)
ds.time
xarray.DataArray
'time'
- time: 248
- array([cftime.DatetimeNoLeap(2005, 1, 15, 1, 30, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(2005, 1, 15, 4, 30, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(2005, 1, 15, 7, 30, 0, 0, has_year_zero=True),
...,
cftime.DatetimeNoLeap(2005, 2, 14, 16, 30, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(2005, 2, 14, 19, 30, 0, 0, has_year_zero=True),
cftime.DatetimeNoLeap(2005, 2, 14, 22, 30, 0, 0, has_year_zero=True)],
dtype=object)
- Coordinates:
- time
(time)
object
2005-01-15 01:30:00 ... 2005-02-...