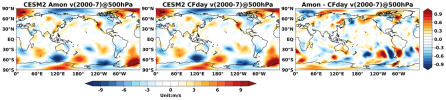
Hello, I would like to ask about the difference between day and CFday in CMIP6, I downloaded the variable va of CESM2 amip experiment from CMIP6 official website, it is r1i1p1f1 table id there are two, one is Amon and the other one is CFday, why I used ncl's vinth2p to interpolate CFday, and then did the monthly average processing to compare with Amon, but the monthly average result of CFday is similar to Amon's distribution, then I did a difference between Amon and CFday, and then I did a difference between Amon and CFday. Why did I use ncl vinth2p to interpolate CFday, and then do the monthly average treatment to compare with Amon, but the monthly average result of CFday and Amon distribution is almost the same, and then I did a difference between Amon and CFday, the result will have to write the deviation of the region, why is this? Or is it that the interpolation method of the output variables in CMIP6 is not the same as vinth2p,This is a picture of mine and the code to run it

Code:
procedure copyVarAttributes(srcVar:numeric, dstVar:numeric)
begin
attrNames = getvaratts(srcVar)
do name=0,dimsizes(attrNames)-1
attrName = attrNames(name)
dstVar@$attrName$ = srcVar@$attrName$
end do
end
folder_path = "/data3/users/2022ghd/sqy/AMIP/CESM2/uvwt/output_year/"
output_path = "/data3/users/2022ghd/sqy/AMIP/CESM2/CESM2_Daily/"
model = "CESM2"
system("if [ ! -d " + output_path + " ]; then mkdir -p " + output_path + "; fi")
p_levs = (/1.0, 3.0, 5.0, 7.0, 10.0, 30.0, 50.0, 70.0, 100.0, 150.0, 200.0, 250.0, 300.0, 350.0, 400.0, 450.0, 500.0,\
550.0, 600.0, 650.0, 700.0, 750.0, 775.0, 800.0, 825.0, 850.0, 875.0, 900.0, 925.0, 950.0, 975.0, 1000.0/)
levels = p_levs
levels@units = "hPa"
levels@long_name = "Pressure Surface"
setfileoption("nc","SuppressClose",False)
do year = 1979, 2014
file_name = systemfunc("ls "+ folder_path + "*" + year + "*.nc")
print("Processing " + file_name)
f = addfile(file_name, "r")
P0mb = f->P0 * 0.01
hyam = f->hyam
hybm = f->hybm
ps = f->PS
V_hybrid = f->V
if (dimsizes(dimsizes(hyam)) .gt. 1) then
hyam_1d = hyam(0,:)
hybm_1d = hybm(0,:)
else
hyam_1d = hyam
hybm_1d = hybm
end if
vonp = vinth2p(V_hybrid, hyam_1d, hybm_1d, levels, ps, 2, P0mb, 1, False)
vnomask = vinth2p(V_hybrid, hyam_1d, hybm_1d, levels, ps, 2, P0mb, 1, True)
nlev = dimsizes(levels)
nlat = dimsizes(f->lat)
nlon = dimsizes(f->lon)
v_all = tofloat(vonp(:,:,::-1,:))
v_nomask = tofloat(vnomask(:,:,::-1,:))
delete(vonp)
delete(vnomask)
copyVarAttributes(V_hybrid, v_all)
copyVarAttributes(V_hybrid, v_nomask)
delete(V_hybrid)
pure_file_name = systemfunc("basename " + file_name)
out_filename = output_path + model + "_AMIP_daily_" + year + ".nc"
system("rm -f " + out_filename)
ncdf = addfile(out_filename, "c")
fAtt = True
fAtt@title = model + " AMIP r1i1p1f1 files interpolated from hybrid to pressure(vinth2p_cam)"
fAtt@source_file = model + " output"
fAtt@conventions = "none"
fAtt@creation_date = systemfunc("date")
fileattdef(ncdf,fAtt)
dimNames = (/"time","level","lat","lon"/)
dimSizes = (/dimsizes(f->time),nlev,nlat,nlon/)
dimUnlim = (/-1,False,False,False/)
filedimdef(ncdf,dimNames,dimSizes,dimUnlim)
filevardef(ncdf,"v",typeof(v_all),(/"time","level","lat","lon"/))
filevardef(ncdf,"v_nomask",typeof(v_nomask),(/"time","level","lat","lon"/))
ncdf->v=(/v_all/)
ncdf->v_nomask=(/v_nomask/)
end do
exit
end