Dr. Oleson,
Thank you for your guidance.
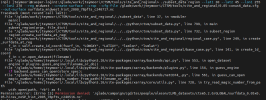
I tried this scripts in both derecho and casper but I face this error:
./subset_data region --lat1 30 --lat2 45 --lon1 255 --lon2 270 --reg midwest --create-surface --crop --cfg-file
Error:
subset_data region: error: argument --cfg-file: expected one argument
1- Do I use the following script? Do you think the Red scripts are true?
cd /CTSM/tools/site_and_regional :
./subset_data region --lat1 30 --lat2 45 --lon1 255 --lon2 270 --reg midwest --create-surface --crop --cfg-file
cd /CTSM/tools/site_and_regional/subset_data_regional:
cp /glade/work/oleson/ctsm5.2.0/tools/site_and_regional/0.05-conus6_data.cfg --out-surface surfdata_midwest_hist_2000_78pfts_c240628.nc landmask.nc
ncrename -v .PFTDATA_MASK,landmask landmask.nc
ncks -A -v .PFTDATA_MASK /glade/work/oleson/ctsm5.2.0/tools/site_and_regional/0.05-conus6_data.cfg --out-surfac surfdata_midwest_hist_2000_78pfts_c240628.nc landmask.nc
ncrename -v .PFTDATA_MASK,mod_lnd_props landmask.nc
ncks --rgr infer --rgr scrip=scrip.nc landmask.nc foo.nc
/glade/u/apps/derecho/23.09/spack/opt/spack/esmf/8.6.0/cray-mpich/8.1.27/oneapi/2023.2.1/7haa/bin/ESMF_Scrip2Unstruct scrip.nc lnd_mesh.nc 0
in the case directory in home : cd midwest
./xmlchange NTASKS=-4
./xmlchange NTASKS_PER_INST=144
./xmlchange JOB_WALLCLOCK_TIME=12:00:00
./xmlchange JOB_QUEUE=main
./xmlchange PIO_TYPENAME=pnetcdf
./xmlchange STOP_OPTION=nyears
./xmlchange STOP_N=1
./xmlchange LND_NX=?
./xmlchange LND_NY=?
./xmlchange ATM_NX=?
./xmlchange ATM_NY=?
./xmlchange RUN_STARTDATE=2012-01-01
./case.setup
fsurdat = '/glade/campaign/cgd/tss/people/oleson/CLM5_datasets/ctsm5.2.0/surfdata_0.05-conus5-nomask-casper-noSP_hist_2000_78pfts_c240516.nc'
Update lines in env_run.xml:
./xmlchange LND_DOMAIN_MESH=/glade/work/jteymoori/CTSM/tools/site_and_regional/subset_data_regional/lnd_mesh.nc
./xmlchange ATM_DOMAIN_MESH=/glade/work/jteymoori/CTSM/tools/site_and_regional/subset_data_regional/lnd_mesh.nc
./xmlchange MASK_MESH=/glade/work/jteymoori/CTSM/tools/site_and_regional/subset_data_regional/lnd_mesh.nc
./case.build
./case.submit
2- What values should be entered for LND_NX, LND_NY, ATM_NX, ATM_NY ؟
Thank you for your guidance.
I tried this scripts in both derecho and casper but I face this error:
./subset_data region --lat1 30 --lat2 45 --lon1 255 --lon2 270 --reg midwest --create-surface --crop --cfg-file
Error:
subset_data region: error: argument --cfg-file: expected one argument
1- Do I use the following script? Do you think the Red scripts are true?
cd /CTSM/tools/site_and_regional :
./subset_data region --lat1 30 --lat2 45 --lon1 255 --lon2 270 --reg midwest --create-surface --crop --cfg-file
cd /CTSM/tools/site_and_regional/subset_data_regional:
cp /glade/work/oleson/ctsm5.2.0/tools/site_and_regional/0.05-conus6_data.cfg --out-surface surfdata_midwest_hist_2000_78pfts_c240628.nc landmask.nc
ncrename -v .PFTDATA_MASK,landmask landmask.nc
ncks -A -v .PFTDATA_MASK /glade/work/oleson/ctsm5.2.0/tools/site_and_regional/0.05-conus6_data.cfg --out-surfac surfdata_midwest_hist_2000_78pfts_c240628.nc landmask.nc
ncrename -v .PFTDATA_MASK,mod_lnd_props landmask.nc
ncks --rgr infer --rgr scrip=scrip.nc landmask.nc foo.nc
/glade/u/apps/derecho/23.09/spack/opt/spack/esmf/8.6.0/cray-mpich/8.1.27/oneapi/2023.2.1/7haa/bin/ESMF_Scrip2Unstruct scrip.nc lnd_mesh.nc 0
in the case directory in home : cd midwest
./xmlchange NTASKS=-4
./xmlchange NTASKS_PER_INST=144
./xmlchange JOB_WALLCLOCK_TIME=12:00:00
./xmlchange JOB_QUEUE=main
./xmlchange PIO_TYPENAME=pnetcdf
./xmlchange STOP_OPTION=nyears
./xmlchange STOP_N=1
./xmlchange LND_NX=?
./xmlchange LND_NY=?
./xmlchange ATM_NX=?
./xmlchange ATM_NY=?
./xmlchange RUN_STARTDATE=2012-01-01
./case.setup
fsurdat = '/glade/campaign/cgd/tss/people/oleson/CLM5_datasets/ctsm5.2.0/surfdata_0.05-conus5-nomask-casper-noSP_hist_2000_78pfts_c240516.nc'
Update lines in env_run.xml:
./xmlchange LND_DOMAIN_MESH=/glade/work/jteymoori/CTSM/tools/site_and_regional/subset_data_regional/lnd_mesh.nc
./xmlchange ATM_DOMAIN_MESH=/glade/work/jteymoori/CTSM/tools/site_and_regional/subset_data_regional/lnd_mesh.nc
./xmlchange MASK_MESH=/glade/work/jteymoori/CTSM/tools/site_and_regional/subset_data_regional/lnd_mesh.nc
./case.build
./case.submit
2- What values should be entered for LND_NX, LND_NY, ATM_NX, ATM_NY ؟